Abstract
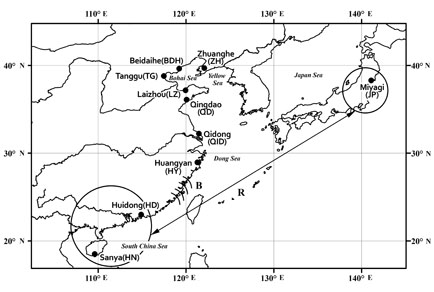
The genetic diversity and structure of 10 populations of Cyclina sinensis distributed along coastal regions in China were inves-tigated by sequencing ribosomal DNA internal transcribed spacer1 (ITS1). The lengths of the ITS1 sequences of C. sinensis ranged from 564 to 595 nucleotides. Forty-two allelic sequences [nucleotide diversity; π=0.033; θ (per site) based on the total number of mutations=0.048] have been identified from a total of 80 individuals. Phylogenetic analysis of these sequences, using a sample from Japan as outgroup, recovered a topology containing two major clades. One clade comprised the samples from the China Bohai Sea, the Yellow Sea and the Dong Sea (northern and middle parts of the China Sea), the other clade rep-resented the those from the South China Sea. FST values indicated significant differences in each pairwise combination of pop-ulations representing each of the two clades, while the AMOVA analysis showed that the majority of genetic variation (67.7%) was attributable to variation between the two main clades, with 25.7% attributable to within-population variation and 6.6% to between populations within groups. These results suggest strong genetic structure among the Chinese populations of C. sinen-sis. Evolutionary rate analysis implies that the two main clades have experienced population isolation since the late Pleistocene (approximately 0.35and1.91MY ago), due to coastal freshwater intrusions and/or cold current upwelling.