Abstract
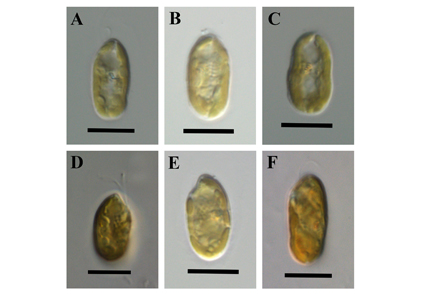
A new species, Cryptomonas indica sp. nov., is described from Western Ghats, India, based on morphological and molecular data. Phylogenetic relationships inferred from nuclear small and large subunit ribosomal DNA, internal transcribed spacer 2, and plastid psbA sequences show that the new species forms a separate lineage on the phylogenetic tree of the genus Cryptomonas. This new species is included in the basal clade of Cryptomonas tree, which consists of undescribed taxa from Europe and South Korea. Cells of this species are up to 19 µm in length, elliptical to slightly asymmetrical in broad view, and have a plastid with four pyrenoids. Cryptomonas indica has been observed in one locality, the Northern region of the Western Ghats. Notably, this is the first Cryptomonas taxon described from the Indian subcontinent with both morphology and molecular information.
References
<p>Byun, Y. & Han, K. (2006) PseudoViewer: web application and webservice for visualizing RNA pseudoknots and secondary structures. <em>Nucleic Acids Research</em> 34: 416–422. https://doi.org/10.1093/nar/gkl210</p>
<p>Choi, B., Son, M., Kim, J.I. & Shin, W. (2013) Taxonomy and phylogeny of the genus <em>Cryptomonas</em> (Cryptophyceae, Cryptophyta) from Korea. <em>Algae </em>28: 307–330. https://doi.org/10.4490/algae.2013.28.4.307</p>
<p>Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. <em>Nature methods</em> 9 (8): 772–772. https://doi.org/10.1038/nmeth.2109</p>
<p>Eddy, S.R. (1998) Profile hidden Markov models. <em>Bioinformatics</em> <em>(Oxford, England)</em> 14 (9): 755–763. https://doi.org/10.1093/bioinformatics/14.9.755</p>
<p>Ehrenberg, C.G. (1831)<em> Symbolae physicae seu icones et descriptiones animalium evertebratorum sepositis insectis quae ex itinere per Africanum Borealem et Asiam Occidentalem Friderici Guilelmi Hemprich et Christiani Godofredi Ehrenberg medicinae et chirurgiae doctorum studio novae aut illustratae redierunt</em>. Berlin, Mittler.</p>
<p>Gadgil, M. & Meher-Homji, V.M. (1990) Ecological diversity. <em>In:</em> Daniels, J.C. & Serrao, J.S. (Eds.) <em>Proceedings of the Centenary Seminar of the Bombay Natural History Society.</em> Bombay Natural History Society and Oxford University Press, Bombay, pp. 175–198.</p>
<p>Gandhi, H.P. (1957) The freshwater diatoms from Radhanagari-Kolhapur. <em>Ceylon Journal of Science (Biology Section)</em> 1: 45–47.</p>
<p>Gandhi, H.P. (1959) Freshwater Diatom flora of the Panhalgarh Hillfort in the Kolhapur district. <em>Hydrobiologia</em> 14: 93–129. https://doi.org/10.1007/BF00042594</p>
<p>Gandhi, H.P. (1999) Fresh-water diatoms of central Gujarat: with a review and some others. Bishen Singh Mahendra Pal Singh, Dehradun, 324 pp.</p>
<p>Gillespie, J.J., Johnston, J.S., Cannone, J.J. & Gutell, R.R. (2006) Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of <em>Apis mellifera</em> (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. <em>Insect Molecular Biology</em> 15: 657–686. https://doi.org/10.1111/j.1365-2583.2006.00689.x</p>
<p>Gusev, E., Podunay, Y., Martynenko, N., Shkurina, N. & Kulikovskiy, M.<strong> (</strong>2020) Taxonomic studies of <em>Cryptomonas lundii</em> clade (Cryptophyta: Cryptophyceae) with description of a new species from Vietnam. <em>Fottea</em> 20 (2): 137–143. https://doi.org/10.5507/fot.2020.004</p>
<p>Hoef-Emden, K. (2007) Revision of the genus <em>Cryptomonas</em> (Cryptophyceae) II: Incongruences between classical morphospecies concept and molecular phylogeny in smaller pyrenoid-less cells. <em>Phycologia</em> 46 (4): 402–428. https://doi.org/10.2216/06-83.1</p>
<p>Hoef-Emden, K. & Melkonian, M. (2003) Revision of the genus <em>Cryptomonas</em> (Cryptophyceae): A combination of molecular phylogeny and morphology provides insights into a long-hidden dimorphism. <em>Protist</em> 154: 371–409. https://doi.org/10.1078/143446103322454130</p>
<p>Hoef-Emden, K. & Archibald, J.M. (2017) Cryptophyta (Cryptomonads).<em> In: </em>Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J. & Corliss, J.O. (eds.) Handbook of the Protists. Springer International Publishing, Cham, Switzerland. https://doi.org/10.1007/978-3-319-28149-0_35</p>
<p>Jindal, R., Thakur, R.K., Singh, U.B. & Ahluwalia, A.S. (2014) Phytoplankton dynamics and species diversity in a shallow eutrophic, natural mid-altitude lake in Himachal Pradesh (India): role of physicochemical factors. <em>Chemistry and Ecology</em> 30 (4): 328–338. https://doi.org/10.1080/02757540.2013.871267</p>
<p>Karthick, B., Kociolek, J.P., Mahesh, M.K. & Ramachandra, T.V. (2011) The diatom genus <em>Gomphonema</em> Ehrenberg in India: Checklist and description of three new species. <em>Nova Hedwigia</em> 93 (1–2): 211–236.</p>
<p>https://doi.org/10.1127/0029-5035/2011/0093-0211</p>
<p>Karthick, B. & Kociolek, J.P. (2012) Reconsideration of the <em>Gomphonema</em> (Bacillariophyceae) species from Kolhapur, Northern Western Ghats, India: Taxonomy, typification and biogeography of the species reported by H. P. Gandhi. <em>Phycological Research</em> 60 (3): 179–198. https://doi.org/10.1111/j.1440-1835.2012.00649.x</p>
<p>Karthick, B., Nautiyal, R., Kociolek, J.P. & Ramachandra, T.V. (2015) Two new species of <em>Gomphonema </em>(Bacillariophyceae) from Doon Valley, Uttarakhand, India. <em>Nova Hedwigia, Beiheft </em>144: 165–174. https://doi.org/10.1127/nova_suppl/2015/0037</p>
<p>Katoh, K. & Toh, H. (2010) Parallelization of the MAFFT multiple sequence alignment program. <em>Bioinformatics</em> 26: 1899–1900. https://doi.org/10.1093/bioinformatics/btq224</p>
<p>Keller, A., Schleicher, T., Schultz, J., Müller, T., Dandekar, T. & Wolf, M. (2009) 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. <em>Gene</em> 430: 50–57. https://doi.org/10.1016/j.gene.2008.10.012</p>
<p>Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. <em>Molecular Biology and Evolution</em> 35: 1547–1549. https://doi.org/10.1093/molbev/msy096</p>
<p>Martynenko, N.A., Gusev, E.S., Kapustin, D.A., Guseva, E.E. & Kulikovskiy, M.S. (2020a) <em>Cryptomonas cattiensis</em> <em>sp. nov.</em> (Cryptophyta: Cryptophyceae), a new species described from Vietnam. <em>Phytotaxa</em> 452 (2): 127–136.</p>
<p>https://doi.org/10.11646/phytotaxa.454.2.4</p>
<p>Martynenko, N.A., Gusev, E.S., Kulizin, P.V., Guseva, E.E., Mccartney, K. & Kulikovskiy, M. S. (2020b) A new species of <em>Cryptomonas</em> (Cryptophyceae) from the Western Urals (Russia). <em>European Journal of Taxonomy</em> 649: 1–12. https://doi.org/10.5852/ejt.2020.649</p>
<p>Mcfadden, G.I. & Melkonian, M. (1986) Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. <em>Phycologia</em> 25: 551–557. https://doi.org/10.2216/i0031-8884-25-4-551.1</p>
<p>Menon, S. & Bawa, K.S. (1997) Applications of geographic information systems, remote-sensing, and a landscape ecology approach to biodiversity conservation in the Western Ghats. <em>Current science</em> 73 (2): 134–145. https://www.jstor.org/stable/24098267</p>
<p>Nakayama, T., Watanabe, S., Mitsui, K., Uchida, H. & Inouye, I. (1996) The phylogenetic relationship between the Chlamydomonadales and Chlorococcales inferred from 18S rDNA sequence data. <em>Phycological research</em> 44 (1): 47–55. https://doi.org/10.1111/j.1440-1835.1996.tb00037.x</p>
<p>Pascal, J.P. (1988) Wet evergreen forests of the Western Ghats of India: ecology, structure, floristic composition, and succession. French Institute, Pondicherry, India.</p>
<p>Patil, S.R., Patil, S.S. & Sathe, T.V. (2014) Status of freshwater bodies from Ajara tahsil of Kolhapur district (MS), India with special reference to morphometric characteristics. <em>IOSR-JESTFT</em> 8 (9): 17–22. https://doi.org/10.9790/2402-08943137</p>
<p>Patidar, S.K., Chokshi, K., George, B., Bhattacharya, S. & Mishra, S. (2015) Dominance of cyanobacterial and cryptophytic assemblage correlated to CDOM at heavy metal contamination sites of Gujarat, India. <em>Environmental monitoring and assessment</em> 187 (1): 4118. https://doi.org/10.1007/s10661-014-4118-6</p>
<p>Playfair, G.J. (1921) Australian freshwater flagellates. <em>Proceedings of the Linnean Society of New South Wales </em>46: 99–146. https://doi.org/10.5962/bhl.part.14004</p>
<p>Pruesse, E., Peplies, J. & Glöckner, F.O. (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. <em>Bioinformatics</em> 28: 1823–1829. https://doi.org/10.1093/bioinformatics/bts252</p>
<p>Radhakrishnan, C., Kulikovskiy, M., Glushchenko, A., Kuznetsova, I., Kociolek, P. & Karthick, B. (2018) <em>Oricymba sagarensis</em> (Gandhi) <em>comb. nov.</em>, an endemic diatom from the Western Ghats, India. <em>Phytotaxa</em> 382 (3): 267–274. https://doi.org/10.11646/phytotaxa.382.3.3</p>
<p>Radhakrishnan, C., Pardhi, S., Kulikovskiy, M., Kociolek, J.P. & Karthick, B. (2020) <em>Navicula watveae</em> <em>sp. nov.</em> (Bacillariophyceae) a new diatom species from the Western Ghats, India. <em>Phytotaxa</em> 433 (1): 20–26. https://doi.org/10.11646/phytotaxa.433.1.3</p>
<p>Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. <em>Bioinformatics</em> 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</p>
<p>Seibel, P.N., Müller, T., Dandekar, T., Schultz, J. & Wolf, M. (2006) 4SALE–a tool for synchronous RNA sequence and secondary structure alignment and editing. <em>BMC Bioinformatics</em> 7 (1): 498. https://doi.org/10.1186/1471-2105-7-498</p>
<p>Shanthala, M., Hosmani, S.P. & Hosetti, B.B. (2009) Diversity of phytoplanktons in a waste stabilization pond at Shimoga Town, Karnataka State, India. <em>Environmental Monitoring and Assessment</em> 151 (1–4): 437–443. https://doi.org/10.1007/s10661-008-0287-5</p>
<p>Sharma, B.K. & Pachuau, L. (2016) Diversity of Phytoplankton of a sub-tropical reservoir of Mizoram, northeast India. <em>International Journal of Aquatic Biology</em> 4 (6): 360–369. https://doi.org/10.22034/ijab.v4i6.195</p>
<p>Roy, S., Radhakrishnan, C., Taylor, J.C., Kulikovskiy, M.S. & Karthick, B. (2020) <em>Encyonema keshrii</em>, <em>sp. nov.</em>: a new diatom species (Cymbellales, Bacillariophyceae) from the Indian subcontinent. <em>Cryptogamie, Algologie</em> 41 (2): 7–17. https://doi.org/10.5252/cryptogamie-algologie2020v41a2</p>
<p>Thompson, J.D., Higgins, D.G. & Gibson, T.J. (1994) CLUSTALW: Improving the sensitivity of progressive multiple sequence through weighing, position-specific gap penalties and weight matrix choice. <em>Nucleic Acids Research</em> 22: 4673–4680. https://doi.org/10.1093/nar/22.22.4673</p>
<p>Vigneshwaran, A., Kulikovskiy, M.S., Glushchenko, A., Kociolek, J.P. & Karthick, B. (2019) A new species of <em>Cymbella</em> (Bacillariophyceae, Cymbellaceae) from the Pavana River, Western Ghats, India. <em>Phytotaxa</em> 395 (3): 209–218. https://doi.org/10.11646/phytotaxa.395.3.5</p>
<p>Wuyts, J., Van De Peer, Y. & De Wachter, R. (2001) Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. <em>Nucleic Acids Research</em> 29: 5017–5028. https://doi.org/10.1093/nar/29.24.5017</p>
<p>Xia, X. & Xie, Z. (2001) DAMBE: software package for data analysis in molecular biology and evolution. <em>Journal of heredity</em> 92 (4): 371–373. https://doi.org/10.1093/jhered/92.4.371</p>
<p>Xia, X., Xie, Z., Salemi, M., Chen, L. & Wang, Y. (2003) An index of substitution saturation and its application. <em>Molecular phylogenetics and evolution</em> 26 (1): 1–7. https://doi.org/10.1016/S1055-7903(02)00326-3</p>
<p>Xia, X. & Lemey, P. (2009) Assessing substitution saturation with DAMBE. <em>The phylogenetic handbook: a practical approach to DNA and protein phylogeny</em> 2: 615–630.</p>
<p>Xia, S., Liu, G.X. & Hu, Z.Y. (2013) Morphological examination and phylogenetic position of two newly recorded freshwater <em>Cryptomonas</em> species (Cryptophyceae) from China. <em>Journal of Systematics and Evolution</em> 51 (2): 212–222. https://doi.org/10.1111/j.1759-6831.2012.00227.x</p>
<p>Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. <em>Nucleic Acids Research</em> 31: 3406–3415. https://doi.org/10.1093/nar/gkg595</p>