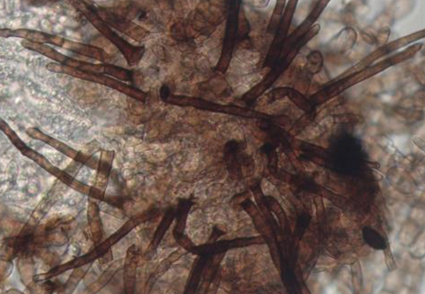
Abstract
Collection of three sexual morphic fungal species occurring on the leaf surface of three plant species (Dalbergia obtusifolia, Syzygium fluviatile and Ziziphus mauritiana) collected in Yunnan Province, China, have shown that they are morphologically distinct. Phylogenetic analyses of partial sequences of internal transcribed spacer region (ITS) and large subunit rDNA (LSU) placed the three species in Trichomerium (Trichomeriaceae) but are sufficiently different from all known species. Therefore, three new species, Trichomerium multisetosum, T. xishuangbannaense and T. yunnanense are described with detailed, illustrated descriptions and notes.
References
<p>Batista, A.C. & Ciferri, R. (1963) Capnodiales. <em>Saccardoa</em> 2: 1–296.</p>
<p>Chomnunti, P., Bhat, D.J., Jones, E.B.G., Chukeatirote, E., Bahkali, A.H. & Hyde, K.D. (2012) <em>Trichomeriaceae</em>, a new sooty mould family of Chaetothyriales. <em>Fungal Diversity</em> 56: 63–76. https://doi.org/10.1007/s13225-012-0197-2</p>
<p>Chomnunti, P., Hongsanan, S., Hudson, B.A., Tian, Q., Peršoh, D., Dhami, M.K., Alias, A.S., Xu, J.C., Liu, X.Z., Stadler, M. & Hyde, K.D. (2014) The sooty moulds. <em>Fungal Diversity</em> 66: 1–36. https://doi.org/10.1007/s13225-014-0278-5</p>
<p>Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. <em>Studies in Mycology</em> 50: 19–22.</p>
<p>Crous, P.W., Wingfield, M.J, Schumacher, R.K., Summerell, B.A., Giraldo, A., Gené, J., Guarro, J., Wanasinghe, D.N., Hyde, K.D., Camporesi, E., Gareth Jones, E.B., Thambugala, K.M., Malysheva, E.F., Malysheva, V.F., Acharya, K., Álvarez, J., Alvarado, P., Assefa, A., Barnes, C.W., Bartlett, J.S., Blanchette, R.A., Burgess, T.I., Carlavilla, J.R., Coetzee, M.P.A., Damm, U., Decock, C.A. den Breeÿen, A., de Vries, B., Dutta, A.K., Holdom, D.G., Latham, S.R., Manjón, J.L., Marincowitz, S., Mirabolfathy, M., Moreno, G., Nakashima, C., Papizadeh, M., Fazeli, S.A.S., Amoozegar, M.A., Romberg, M.K., Shivas, R.G., Stalpers, J.A., Stielow, B., Stukely, M.J.C., Swart, W.J., Tan, Y.P., van der Bank, M., Wood, A.R., Zhang, Y. & Groenewald, J.Z. (2015) Fungal Planet Description Sheets: 281–319. <em>Persoonia</em> 33: 212−289. https://doi.org/10.3767/003158514X685680</p>
<p>Crous, P.W., Wingfield, M.J., Burgess, T.I. <em>et al.</em> (2017) Fungal Planet description sheets: 625–715. <em>Persoonia</em> 39: 270–467. https://doi.org/10.3767/persoonia.2017.39.11</p>
<p>Dissanayake, a.J., Bhunjun, C.S., Maharachchikumbura, S.S.N. & Liu, J.K. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. <em>Mycosphere </em>11: 2653–2677. https://doi.org/10.5943/mycosphere/11/1/18</p>
<p>Felsenstein, J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. <em>Evolution</em> 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x</p>
<p>Gueidan, C., Aptroot, A., da Silva, M.E., Badali, H. & Stenroos, S. (2014) A reappraisal of orders and families within the subclass Chaetothyriomycetidae (Eurotiomycetes, Ascomycota). <em>Mycological Progress</em> 13: 1027–1039. https://doi.org/10.1007/s11557-014-0990-2</p>
<p>Hongsanan, S., Tian, Q., Hyde, K.D. & Hu, D.M. (2016) The asexual morph of <em>Trichomerium</em> <em>gloeosporum</em>. <em>Mycosphere</em> 7: 1473–1479. https://doi.org/10.5943/mycosphere/7/9/18</p>
<p>Hyde, K.D., Hongsanan, S., Jeewon, R., <em>et al.</em> (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. <em>Fungal Diversity</em> 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x</p>
<p>Index Fungorum (2021) Available from: http://www.indexfungorum.org/Names/Names.asp. (Accessed July 2021)</p>
<p>Kirk, P.M., Cannon, P.F., Minter, D.W. & Stalpers, J.A. (2008) Dictionary of the Fungi. (10thedn). Wallingford, UK.</p>
<p>Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. <em>Molecular Biology and Evolution</em> 33: 1870–1874. https://doi.org/10.1093/molbev/msw054</p>
<p>Liu, X.Y., Udayanga, D., Luo, Z.L., Chen, L.J., Zhou, D.Q., Su, H.Y. & Hyde, K.D. (2015) Backbone tree for Chaetothyriales with four new species of <em>Minimelanolocus</em> from aquatic habitats. <em>Fungal Biology</em> 119: 1046–1062. https://doi.org/10.1016/j.funbio.2015.08.005</p>
<p>Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. <em>In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond.</em> pp. 1–8. https://doi.org/10.1145/2335755.2335836</p>
<p>Nascimento, M.M.F., Selbmannc, L., Sharifyniab, S., Al-Hatmib, A.M.S., Voglmayr, H., Vicente, V.A., Deng, S.W., Kargl, A., Moussa, T.A.A., Al-Zahrani, H.S., Almaghrabi, O.A. & de Hoog, G.S. (2016) <em>Arthrocladium</em>, an unexpected human opportunist in <em>Trichomeriaceae</em> (Chaetothyriales). <em>Fungal Biology</em> 120: 207–218. https://doi.org/10.1016/j.funbio.2015.08.018</p>
<p>Maharachchikumbura, S.S.N., Haituk, S., Pakdeeniti, P., Al-Sadi, A.M., Hongsanan, S., Chomnunti, P. & Cheewangkoon, R. (2018) <em>Phaeosaccardinula</em> <em>coffeicola</em> and <em>Trichomerium chiangmaiensis</em>, two new species of Chaetothyriales (Eurotiomycetes) from Thailand. <em>Mycosphere</em> 9: 769–778. https://doi.org/10.5943/mycosphere/9/4/5</p>
<p>Phillips, A.J.L., Alves, A., Pennycook, S.R., Johnston, P.R., Ramaley, A., Akulov, A. & Crous, P.W. (2008) Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. <em>Persoonia</em> 21: 29–55. https://doi.org/10.3767/003158508X340742</p>
<p>Réblová, M., Untereiner, W.A. & Réblová, K. (2013) Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multi-gene phylogenetic analyses and comparison of ITS secondary structure. <em>PLoS ONE</em> 8: e63547. https://doi.org/10.1371/journal.pone.0063547</p>
<p>Spegazzini, C. (1918) Notas micológicas. <em>Physis Revista de la Sociedad Argentina de Ciencias Naturales</em> 4: 281–295.</p>
<p>Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. <em>Bioinformatics</em> 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</p>
<p>Sun, W., Su, L., Yang, S., Sun, J., Liu, B., Fu, R., Wu, B., Liu, X., Cai, L., Guo, L. & Xiang, M. (2020) Unveiling the Hidden Diversity of Rock-Inhabiting Fungi: Chaetothyriales from China. <em>Journal of Fungi</em> 6: 187. https://doi.org/10.3390/jof6040187</p>
<p>Swofford, D.L. (2002) PAUP: Phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associates, Sunderland.</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990</p>
<p>White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.<em> In: </em>Innis MA, Gelfand, D.H., Sninsky, J.J. & White, T.J. (eds.) <em>PCR protocols: a guide to methods and applications.</em> San Diego: Academic Press, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T., Tedersoo, L., Rajeshkumar, K.C., Zhao, R.L., Aptroot, A., Leontyev, D.V., Saxena, R.K., Tokarev, Y.S., Dai, D.Q., Letcher, P.M., Stephenson, S.L., Ertz, D., Lumbsch, H.T., Haelewaters, D., Bensch, K., Madrid, H., Kukwa, M., Issi, I.V., Lateef, A.A., Phillips, A.J.L., Selbmann, L., Pfiegler, W.P., Horváth, E., Pereira, O.L., Kirk, P.M., Kolaříková, Z., Raja, H.A., Radek, R., Papp, V., Dima, B., Ma, J., Malosso, E., Takamatsu, S., Rambold, G., Gannibal, P.B., Triebel, D., Gautam, A.K., Avasthi, S., Suetrong, S., Timdal, E., Fryar, S.C., Delgado, G., Réblová, M., Doilom, M., Dolatabadi, S., Pawłowska, J., Humber, R.A., Kodsueb, R., Sánchez-Castro, I., Goto, B.T., Silva, D.K.A., de Souza, F.A., Oehl, F., Santos da Silva, G.A., Silva, I.R., Błaszkowski, J., Jobim, K., Maia, L.C., Barbosa, F.R., Fiuza, P.O., Divakar, P.K., Shenoy, B.D., Castañeda-Ruiz, R.F., Somrithipo, S., Karunarathna, S.C., Tibpromma, S., Mortimer, P.E., Wanasinghe, D.N., Phookamsak, R., Xu, J.C., Wang, Y., Fenghua, T., Alvarado, P., Li, D.W., Kušan, I., Matočec, N., Maharachchikumbura, S.S.N., Papizadeh, M., Heredia, G., Wartchow, F., Bakhshi, M., Boehm, E., Youssef, N., Hustad, V.P., Lawrey, J.D., Santiago, ALCMA, Bezerra, J.D.P., Souza-Motta, C.M., Firmino, A.L., Tian, Q., Houbraken, J., Hongsanan, S., Tanaka, K., Dissanayake, A.J., Monteiro, J.S., Grossart, H.P., Suija, A., Weerakoon, G., Etayo, J., Tsurykau, A., Kuhnert, E., Vázquez, V., Mungai, P., Damm, U., Li, Q.R., Zhang, H., Boonmee, S., Lu, Y.Z., Becerra, A.G., Kendrick, B., Brearley, F.Q., Motiejűnaitë, J., Sharma, B., Khare, R., Gaikwad, S., Wijesundara, D.S.A., Tang, L.Z., He, M.Q., Flakus, A., RodriguezFlakus, P., Zhurbenko, M.P., McKenzie, E.H.C., Stadler, M., Bhat, D.J., Liu, J.K., Raza, M., Jeewon, R., Nassonova, E.S., Prieto, M., Jayalal, R.G.U., Yurkov, A., Schnittler, M., Shchepin, O.N., Novozhilov, Y.K., Liu, P., Cavender, J.C., Kang, Y.Q., Mohammad, S., Zhang, L.F., Xu, R.F., Li, Y.M., Dayarathne, M.C., Ekanayaka, A.H., Wen, T.C., Deng, C.Y. & Navathe, S. (2020) Outline of Fungi and fungus-like taxa. <em>Mycosphere</em> 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8</p>
<p>Chomnunti, P., Bhat, D.J., Jones, E.B.G., Chukeatirote, E., Bahkali, A.H. & Hyde, K.D. (2012) <em>Trichomeriaceae</em>, a new sooty mould family of Chaetothyriales. <em>Fungal Diversity</em> 56: 63–76. https://doi.org/10.1007/s13225-012-0197-2</p>
<p>Chomnunti, P., Hongsanan, S., Hudson, B.A., Tian, Q., Peršoh, D., Dhami, M.K., Alias, A.S., Xu, J.C., Liu, X.Z., Stadler, M. & Hyde, K.D. (2014) The sooty moulds. <em>Fungal Diversity</em> 66: 1–36. https://doi.org/10.1007/s13225-014-0278-5</p>
<p>Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. <em>Studies in Mycology</em> 50: 19–22.</p>
<p>Crous, P.W., Wingfield, M.J, Schumacher, R.K., Summerell, B.A., Giraldo, A., Gené, J., Guarro, J., Wanasinghe, D.N., Hyde, K.D., Camporesi, E., Gareth Jones, E.B., Thambugala, K.M., Malysheva, E.F., Malysheva, V.F., Acharya, K., Álvarez, J., Alvarado, P., Assefa, A., Barnes, C.W., Bartlett, J.S., Blanchette, R.A., Burgess, T.I., Carlavilla, J.R., Coetzee, M.P.A., Damm, U., Decock, C.A. den Breeÿen, A., de Vries, B., Dutta, A.K., Holdom, D.G., Latham, S.R., Manjón, J.L., Marincowitz, S., Mirabolfathy, M., Moreno, G., Nakashima, C., Papizadeh, M., Fazeli, S.A.S., Amoozegar, M.A., Romberg, M.K., Shivas, R.G., Stalpers, J.A., Stielow, B., Stukely, M.J.C., Swart, W.J., Tan, Y.P., van der Bank, M., Wood, A.R., Zhang, Y. & Groenewald, J.Z. (2015) Fungal Planet Description Sheets: 281–319. <em>Persoonia</em> 33: 212−289. https://doi.org/10.3767/003158514X685680</p>
<p>Crous, P.W., Wingfield, M.J., Burgess, T.I. <em>et al.</em> (2017) Fungal Planet description sheets: 625–715. <em>Persoonia</em> 39: 270–467. https://doi.org/10.3767/persoonia.2017.39.11</p>
<p>Dissanayake, a.J., Bhunjun, C.S., Maharachchikumbura, S.S.N. & Liu, J.K. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. <em>Mycosphere </em>11: 2653–2677. https://doi.org/10.5943/mycosphere/11/1/18</p>
<p>Felsenstein, J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. <em>Evolution</em> 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x</p>
<p>Gueidan, C., Aptroot, A., da Silva, M.E., Badali, H. & Stenroos, S. (2014) A reappraisal of orders and families within the subclass Chaetothyriomycetidae (Eurotiomycetes, Ascomycota). <em>Mycological Progress</em> 13: 1027–1039. https://doi.org/10.1007/s11557-014-0990-2</p>
<p>Hongsanan, S., Tian, Q., Hyde, K.D. & Hu, D.M. (2016) The asexual morph of <em>Trichomerium</em> <em>gloeosporum</em>. <em>Mycosphere</em> 7: 1473–1479. https://doi.org/10.5943/mycosphere/7/9/18</p>
<p>Hyde, K.D., Hongsanan, S., Jeewon, R., <em>et al.</em> (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. <em>Fungal Diversity</em> 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x</p>
<p>Index Fungorum (2021) Available from: http://www.indexfungorum.org/Names/Names.asp. (Accessed July 2021)</p>
<p>Kirk, P.M., Cannon, P.F., Minter, D.W. & Stalpers, J.A. (2008) Dictionary of the Fungi. (10thedn). Wallingford, UK.</p>
<p>Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. <em>Molecular Biology and Evolution</em> 33: 1870–1874. https://doi.org/10.1093/molbev/msw054</p>
<p>Liu, X.Y., Udayanga, D., Luo, Z.L., Chen, L.J., Zhou, D.Q., Su, H.Y. & Hyde, K.D. (2015) Backbone tree for Chaetothyriales with four new species of <em>Minimelanolocus</em> from aquatic habitats. <em>Fungal Biology</em> 119: 1046–1062. https://doi.org/10.1016/j.funbio.2015.08.005</p>
<p>Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. <em>In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond.</em> pp. 1–8. https://doi.org/10.1145/2335755.2335836</p>
<p>Nascimento, M.M.F., Selbmannc, L., Sharifyniab, S., Al-Hatmib, A.M.S., Voglmayr, H., Vicente, V.A., Deng, S.W., Kargl, A., Moussa, T.A.A., Al-Zahrani, H.S., Almaghrabi, O.A. & de Hoog, G.S. (2016) <em>Arthrocladium</em>, an unexpected human opportunist in <em>Trichomeriaceae</em> (Chaetothyriales). <em>Fungal Biology</em> 120: 207–218. https://doi.org/10.1016/j.funbio.2015.08.018</p>
<p>Maharachchikumbura, S.S.N., Haituk, S., Pakdeeniti, P., Al-Sadi, A.M., Hongsanan, S., Chomnunti, P. & Cheewangkoon, R. (2018) <em>Phaeosaccardinula</em> <em>coffeicola</em> and <em>Trichomerium chiangmaiensis</em>, two new species of Chaetothyriales (Eurotiomycetes) from Thailand. <em>Mycosphere</em> 9: 769–778. https://doi.org/10.5943/mycosphere/9/4/5</p>
<p>Phillips, A.J.L., Alves, A., Pennycook, S.R., Johnston, P.R., Ramaley, A., Akulov, A. & Crous, P.W. (2008) Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. <em>Persoonia</em> 21: 29–55. https://doi.org/10.3767/003158508X340742</p>
<p>Réblová, M., Untereiner, W.A. & Réblová, K. (2013) Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multi-gene phylogenetic analyses and comparison of ITS secondary structure. <em>PLoS ONE</em> 8: e63547. https://doi.org/10.1371/journal.pone.0063547</p>
<p>Spegazzini, C. (1918) Notas micológicas. <em>Physis Revista de la Sociedad Argentina de Ciencias Naturales</em> 4: 281–295.</p>
<p>Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. <em>Bioinformatics</em> 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</p>
<p>Sun, W., Su, L., Yang, S., Sun, J., Liu, B., Fu, R., Wu, B., Liu, X., Cai, L., Guo, L. & Xiang, M. (2020) Unveiling the Hidden Diversity of Rock-Inhabiting Fungi: Chaetothyriales from China. <em>Journal of Fungi</em> 6: 187. https://doi.org/10.3390/jof6040187</p>
<p>Swofford, D.L. (2002) PAUP: Phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associates, Sunderland.</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990</p>
<p>White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.<em> In: </em>Innis MA, Gelfand, D.H., Sninsky, J.J. & White, T.J. (eds.) <em>PCR protocols: a guide to methods and applications.</em> San Diego: Academic Press, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T., Tedersoo, L., Rajeshkumar, K.C., Zhao, R.L., Aptroot, A., Leontyev, D.V., Saxena, R.K., Tokarev, Y.S., Dai, D.Q., Letcher, P.M., Stephenson, S.L., Ertz, D., Lumbsch, H.T., Haelewaters, D., Bensch, K., Madrid, H., Kukwa, M., Issi, I.V., Lateef, A.A., Phillips, A.J.L., Selbmann, L., Pfiegler, W.P., Horváth, E., Pereira, O.L., Kirk, P.M., Kolaříková, Z., Raja, H.A., Radek, R., Papp, V., Dima, B., Ma, J., Malosso, E., Takamatsu, S., Rambold, G., Gannibal, P.B., Triebel, D., Gautam, A.K., Avasthi, S., Suetrong, S., Timdal, E., Fryar, S.C., Delgado, G., Réblová, M., Doilom, M., Dolatabadi, S., Pawłowska, J., Humber, R.A., Kodsueb, R., Sánchez-Castro, I., Goto, B.T., Silva, D.K.A., de Souza, F.A., Oehl, F., Santos da Silva, G.A., Silva, I.R., Błaszkowski, J., Jobim, K., Maia, L.C., Barbosa, F.R., Fiuza, P.O., Divakar, P.K., Shenoy, B.D., Castañeda-Ruiz, R.F., Somrithipo, S., Karunarathna, S.C., Tibpromma, S., Mortimer, P.E., Wanasinghe, D.N., Phookamsak, R., Xu, J.C., Wang, Y., Fenghua, T., Alvarado, P., Li, D.W., Kušan, I., Matočec, N., Maharachchikumbura, S.S.N., Papizadeh, M., Heredia, G., Wartchow, F., Bakhshi, M., Boehm, E., Youssef, N., Hustad, V.P., Lawrey, J.D., Santiago, ALCMA, Bezerra, J.D.P., Souza-Motta, C.M., Firmino, A.L., Tian, Q., Houbraken, J., Hongsanan, S., Tanaka, K., Dissanayake, A.J., Monteiro, J.S., Grossart, H.P., Suija, A., Weerakoon, G., Etayo, J., Tsurykau, A., Kuhnert, E., Vázquez, V., Mungai, P., Damm, U., Li, Q.R., Zhang, H., Boonmee, S., Lu, Y.Z., Becerra, A.G., Kendrick, B., Brearley, F.Q., Motiejűnaitë, J., Sharma, B., Khare, R., Gaikwad, S., Wijesundara, D.S.A., Tang, L.Z., He, M.Q., Flakus, A., RodriguezFlakus, P., Zhurbenko, M.P., McKenzie, E.H.C., Stadler, M., Bhat, D.J., Liu, J.K., Raza, M., Jeewon, R., Nassonova, E.S., Prieto, M., Jayalal, R.G.U., Yurkov, A., Schnittler, M., Shchepin, O.N., Novozhilov, Y.K., Liu, P., Cavender, J.C., Kang, Y.Q., Mohammad, S., Zhang, L.F., Xu, R.F., Li, Y.M., Dayarathne, M.C., Ekanayaka, A.H., Wen, T.C., Deng, C.Y. & Navathe, S. (2020) Outline of Fungi and fungus-like taxa. <em>Mycosphere</em> 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8</p>