Abstract
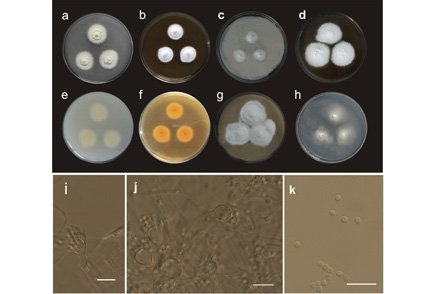
A novel species in Aspergillus section Fumigati is described as Aspergillus sibiricus. The new species was isolated from an open coal mine in Altai, Siberia, Russia and is represented by ex-type strain CBS 143307. We used a polyphasic taxonomic approach to characterise the species. A description based on morphological features is provided and shows that the new species differs morphologically from closely related species (A. assulatus, A. nishimurae and A. waksmanii). Partial sequences of the loci encoding β-tubulin (BenA), calmodulin (CaM), ribosomal polymerase II second largest subunit (RPB2) and internal transcribed spacer rDNA region (ITS1-5.8S-ITS2) were analyzed as well. Sequences data, macro- and micromorphological and physiological characteristics distinguish A. sibiricus from all known species in section Fumigati, series Unilaterales. A. sibiricus, isolated from an acidic habitat (pH 3.2–3.4), is able to grow at pH 2 and is considered an acid-resistant species.
References
<p>Ajello, L. (1968) A taxonomic review of the dermatophytes and related species. <em>Sabouraudia</em> 6: 147–149.</p>
<p>Alcazar-Fuoli, L., Mellado, E., Alastruey-Izquierdo, A., Cuenca-Estrella, M. & Rodriguez-Tudela, J.L. (2008) <em>Aspergillus</em> section <em>Fumigati</em>: antifungal susceptibility patterns and sequence-based identification. <em>Antimicrobial Agents and Chemotherapy </em>52: 1244–1251. https://doi.org/10.1128/AAC.00942-07</p>
<p>Amaral Zettler, L.A., Gómez, F., Zettler, E., Keenan, B.G., Amils, R. & Sogin, M.L. (2002) Microbiology: eukaryotic diversity in Spain’s river of fire. <em>Nature </em>417: 137.</p>
<p>Baker, B.J., Lutz, M.A., Dawson, S.C., Bond, P.L. & Banfield, J.F. (2004) Metabolically active eukaryotic communities in extremely acidic mine drainage. <em>Applied and Environmental Microbiology </em>70: 6264–6271. https://doi.org/10.1128/AEM.70.10.6264-6271.2004</p>
<p>Balajee, S.A., Nickle, D., Varga, J. & Marr, K.A. (2006) Molecular studies reveal frequent misidentification of <em>Aspergillus fumigatus</em> by morphotyping. <em>Eukaryotic Cell </em>5: 1705–1712. https://doi.org/10.1128/EC.00162-06</p>
<p>Banks, D., Burke, S.P. & Gray, C.G. (1997) Hydrogeochemistry of coal mine drainage and other ferruginous waters in north Derbyshire and south Yorkshire, UK. <em>Quarterly Journal of Engineering Geology & Hydrogeology </em>30: 257–280. http://dx.doi.org/10.1144/GSL.QJEG.1997.030.P3.07</p>
<p>Berkeley, M.J. (1860) <em>Outlines of British Fungology</em>. 442 pp.</p>
<p>Fresenius, G. (1863) <em>Beiträge zur Mykologie</em> 3.</p>
<p>Glass, N.L. & Donaldson, G.C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. <em>Applied and Environmental Microbiology </em>61: 1323–1330.</p>
<p>Gross, S. & Robbins, E.I. (2000) Acidophilic and acid-tolerant fungi and yeasts. <em>Hydrobiologia </em>433: 91–109. https://doi.org/10.1023/A:1004014603333</p>
<p>Hong, S.-B., Go, S.-J., Shin, H.-D., Frisvad, J.C. & Samson, R.A. (2005) Polyphasic taxonomy of <em>Aspergillus fumigatus</em> and related species. <em>Mycologia</em> 97: 1316–1329.</p>
<p>Hong, S.-B., Shin, H.-D., Hong, J., Frisvad, J.C., Nielsen, P.V., Varga, J. & Samson, R.A. (2008) New taxa of <em>Neosartorya</em> and <em>Aspergillus</em> in <em>Aspergillus</em> section <em>Fumigati. Antonie van Leeuwenhoek</em> 93: 87–98. https://doi.org/10.1007/s10482-007-9183-1</p>
<p>Houbraken, J. & Samson, R.A. (2011) Phylogeny of <em>Penicillium</em> and the segregation of <em>Trichocomaceae</em> into three families. <em>Studies in Mycology</em> 70: 1–51. https://doi.org/10.3114/sim.2011.70.01</p>
<p>Houbraken, J., Weig, M., Gro, U., Meijer, M. & Bader, O. (2016) <em>Aspergillus oerlinghausenensis</em>, a new mould species closely related to <em>A. fumigatus</em>. <em>FEMS Microbiology Letters</em> 363: fnv236. https://doi.org/10.1093/femsle/fnv236</p>
<p>Houbraken, J., Kocsubé, S., Visagie, C.M., Yilmaz, N. Wang, X.-C., Meijer, M., Kraak, B., Hubka, V., Bensch, K., Samson, R.A. & Frisvad, J.C. (2020) Classification of <em>Aspergillus</em>, <em>Penicillium</em>, <em>Talaromyces</em> and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. <em>Studies in Mycology</em> 95: 5–169. https://doi.org/10.1016/j.simyco.2020.05.002</p>
<p>Hubka, V., Peterson, S.W., Frisvad, J.C., Yaguchi, T., Kubatova, A. & Kolarýk, M. (2013) <em>Aspergillus waksmanii</em> <em>sp. nov.</em> and <em>Aspergillus marvanovae</em> <em>sp. nov.</em>, two closely related species in section <em>Fumigati</em>. <em>International Journal of Systematic and Evolutionary Microbiology</em> 63: 783–789. https://doi.org/10.1099/ijs.0.047076-0</p>
<p>Hubka, V., Dudová, Z., Kubátová, A., Frisvad, J.C., Yaguchi, T., Horie, Y., Jurjević, Ž., Hong, S.-B. & Kolařík, M. (2017) Taxonomic novelties in <em>Aspergillus</em> section <em>Fumigati</em>: <em>A. tasmanicus</em> <em>sp. nov.</em>, induction of sexual state in <em>A. turcosus</em> and overview of related species. <em>Plant Systematics and Evolution </em>303: 787–806. https://doi.org/10.1007/s00606-017-1400-4</p>
<p>Hujslová, M., Kubátová, A., Kostovčík, M. & Kolařík, M. (2012) <em>Acidiella bohemica</em> <em>gen. et sp. nov.</em> and <em>Acidomyces</em> spp. (<em>Teratosphaeriaceae</em>), the indigenous inhabitants of extremely acidic soils in Europe. <em>Fungal Diversity</em> 58: 33–45. http://dx.doi.org/10.1007/s13225-012-0176-7</p>
<p>Hujslová, M., Kubátová, A., Kostovčík, M., Blanchette, R.A., de Beer, Z.W., Chudíčková, M. & Kolařík, M. (2014) Three new genera of fungi from extremely acidic soils. <em>Mycological Progress </em>13: 819–831. http://dx.doi.org/10.1007/s11557-014-0965-3</p>
<p>Hujslová, M., Alena, K., Petra, B., Milada, C. & Miroslav, K. (2017) Extremely acidic soils are dominated by species-poor and highly specific fungal communities. <em>Microbial Ecology </em>73: 321–337. http://dx.doi.org/10.1007/s00248-016-0860-3</p>
<p>Kelly, K.L. (1964) <em>Inter-Society Color Council – National Bureau of Standards Color Name Charts Illustrated with Centroid Colors</em>. Government Printing Office, Washington, DC: US.</p>
<p>Kolařík, M., Hujslová, M. & Vazquéz-Campos, X. (2015) Acidotolerant genus <em>Fodinomyces</em> (<em>Ascomycota: Capnodiales</em>) is a synonym of <em>Acidiell</em>a. <em>Czech Mycology </em>67: 37–38.</p>
<p>Kontoyiannis, D.P. & Bodey, G.P. (2002) Invasive aspergillosis in 2002: an update. <em>European Journal of Clinical Microbiology & Infectious Diseases</em> 21: 161–172. https://doi.org/10.1007/s10096-002-0699-z</p>
<p>Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. <em>Molecular Biology and Evolution </em>35: 1547–1549. https://doi.org/10.1093/molbev/msy096</p>
<p>Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. & Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. <em>Bioinformatics</em> 23: 2947–2948.</p>
<p>Matsuzawa, T., Takaki, G.M.C., Yaguchi, T., Okada, K., Gonoi, T. & Horie, Y. (2014) Two new species of <em>Aspergillus </em>section <em>Fumigati </em>isolated from caatinga soil in the state of Pernambuco, Brazil. <em>Mycoscience</em> 55: 79–88. https://doi.org/10.1016/j.myc.2013.04.001</p>
<p>McLennan, E.I., Ducker, S.C. & Thrower, L.B. (1954) New soil fungi from Australian heathland: <em>Aspergillus</em>, <em>Penicillium</em>, <em>Spegazzinia</em>. <em>Australian Journal of Botany</em> 2 (3): 355–364.</p>
<p>McNeill, J., Barrie, F.R., Buck, W.R., Demoulin, V., Greuter, W., Hawksworth, D., Herendeen, P.S., Knapp, S., Marhold, K., Prado, J., Prud’Homme van Reine, W.F., Smith, G.F., Wiersema, J.H. & Turland, N.J. (2012) <em>International Code of Nomenclature for algae, fungi, and plants (Melbourne Code).</em> Koeltz Scientific Books, Königstein, 208 pp.</p>
<p>Nováková, A., Hubka, V., Saiz-Jimenez, C. & Kolarik, M. (2012) <em>Aspergillus baeticus</em> <em>sp. nov.</em> and <em>Aspergillus thesauricus</em> <em>sp. nov.</em>, two species in section <em>Usti</em> from Spanish caves. <em>International Journal of Systematic and Evolutionary Microbiology</em> 62: 2778–2785. https://doi.org/10.1099/ijs.0.041004-0</p>
<p>Qureshi, A., Maurice, C. & Öhlander, B. (2016) Potential of coal mine waste rock for generating acid mine drainage. <em>Journal of Geochemical Exploration </em>160: 44–54. https://doi.org/10.1016/j.gexplo.2015.10.014</p>
<p>Raper, K.B. & Fennell, D.I. (1965) <em>The genus Aspergillus</em>. Williams & Wilkins, Baltimore, 686 pp.</p>
<p>Saluja, P. & Prasad, G.S. (2007) <em>Debaryomyces singareniensis</em> <em>sp. nov.</em>, a novel yeast species isolated from a coalmine soil in India. <em>FEMS Yeast Research </em>7: 482–488. https://doi.org/10.1111/j.1567-1364.2006.00182.x</p>
<p>Saluja, P., Yelchuri, R.K., Sohal, S.K., Bhagat, G., Paramjit & Prasad, G.S. (2012) <em>Torulaspora indica</em> a novel yeast species isolated from coal mine soils. <em>Antonie van Leeuwenhoek</em> 101: 733–742. https://doi.org/10.1007/s10482-011-9687-6</p>
<p>Samson, R.A., Hong, S.-B., Peterson, S.W., Frisvad, J.C. & Varga, J. (2007) Polyphasic taxonomy of <em>Aspergillus </em>section <em>Fumigati </em>and its teleomorph <em>Neosartorya</em>. <em>Studies in Mycology </em>59: 147–207. https://doi.org/10.3114/sim.2007.59.14</p>
<p>Samson, R.A., Houbraken, J., Thrane, U., Frisvad, J.C. & Andersen, B. (2010) <em>Food and indoor fungi, CBS Laboratory Manual Series 2</em>. CBS-Fungal Biodiversity Centre, Utrecht, 390 pp.</p>
<p>Samson, R.A., Visagie, C.M., Houbraken, J., Hong, S.B., Hubka, V., Klaassen, C.H.W., Perrone, G., Seifert, K.A., Susca, A., Tanney, J.B., Varga, J., Kocsubé, S., Szigeti, G., Yaguchi, T. & Frisvad, J.C. (2014) Phylogeny, identification and nomenclature of the genus <em>Aspergillus</em>. <em>Studies in Mycology</em> 78: 141–173. https://doi.org/10.1016/j.simyco.2014.07.004</p>
<p>Schoch, C.L., Seifert, K.A., Huhndorf, S., Robert, V., Spogue, J.L., Levesque, C.A. & Chen, W. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. <em>Proceedings of the National Academy of Sciences</em> 109: 6241–6246. https://doi.org/10.1073/pnas.1117018109</p>
<p>Selbmann, L., de Hoog, G.S., Zucconi, L., Isola, D., Ruisi, S., Gerrits, A.H.G., Ende, V.A., Ruibal, C., De Leo, F., Urzì, C. & Onofri, S. (2008) Drought meets acid: three new genera in a dothidealean clade of extremotolerant fungi. <em>Studies in Mycology</em> 61: 1–20. https://doi.org/10.3114/sim.2008.61.01</p>
<p>Sugui, J.A., Peterson, S.W., Figat, A., Hansen, B., Samson, R.A., Mellado, E., Cuenca-Estrella, M. & Kwon-Chung, K.J. (2014) Genetic relatedness versus biological compatibility between <em>Aspergillus fumigatus</em> and related species. <em>Journal of Clinical Microbiology</em> 52: 3707–3721. https://doi.org/10.1128/JCM.01704-14</p>
<p>Takada, M., Horie, Y. & Abliz, P. (2001) Two new heterothallic <em>Neosartorya</em> from African soil. <em>Mycoscience</em> 42: 361–367</p>
<p>Tamura, K. & Nei, M. (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. <em>Molecular Biology and Evolution </em>10: 512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023</p>
<p>Udagawa, S., Tsubouchi, H. & Horie, Y. (1991) <em>Neosartorya hiratsukae</em>, a new species of food-borne Ascomycetes. <em>Transactions of the Mycological Society of Japan</em> 32: 23–29.</p>
<p>Ulfig, K. & Korcz, M. (1995) Isolation of keratinolytic fungi from a coal mine dump. <em>Mycopathologia</em> 129: 83–86.</p>
<p>White, T.J., Bruns, T.D., Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>In:</em> Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) <em>PCR protocols: a guide to methods and applications</em>. Academic Press, London, UK, pp. 315–322.</p>
<p>Younger, P.L. (2004) Environmental impacts of coal mining and associated wastes: a geochemical perspective. <em>Geological Society London Special Publications</em> 236: 169–209. https://doi.org/10.1144/GSL.SP.2004.236.01.12</p>
<p>Zak, J.C. & Wildman, H.G. (2004) Fungi in stressful environments. <em>In</em>: Mueller, G.M., Bills, G.F. & Foster, M.S. (Eds.) <em>Biodiversity of fungi, inventory and monitoring methods</em>. Elsevier Academic Press, Amsterdam etc., pp. 303–331.</p>
<p>Zirnstein, I., Arnold, T., Krawczyk-Bärsch, E., Jenk, U., Bernhard, G. & Röske, I. (2012) Eukaryotic life in biofilms formed in a uranium mine. <em>The Open Microbiology Journal </em>1: 83–94. https://doi.org/10.1002/mbo3.17</p>