Abstract
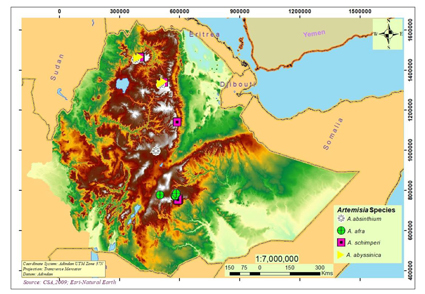
There are four species of Artemisia known in Ethiopia, namely A. schimperi (endemic), A. abyssinica, A. afra and A. absinthium (syn. A. rehan). All these species occurred naturally except A. absinthium, which usually grows in private gardens. The main objective of this study was to reconstruct the molecular phylogeny to determine phylogenetic positions of Ethiopian Artemisia species using the external and internal transcribed spacers (ETS, ITS). A total of 57 sequences (nine new and 48 retrieved from GenBank) were used in maximum parsimony (MP) and maximum likelihood (ML) analyses. The Ethiopian Artemisia species were recovered in three clades corresponding to subgenera (Artemisia, Absinthium and Pacifica). The sequences of Ethiopian A. absinthium (previously known as A. rehan) grouped with A. judaica and are distantly related to the European A. absinthium. Artemisia abyssinica and A. schimperi have affinities to A. subg. Pacifica with adaptations to a relatively cool and dry subalpine tropical climate.
References
<p>Abebe, T., Teshome, B., Abatneh, E. & Yimama, A. (2019) Chemical Composition, Antimicrobial Activity and Antioxidant Property of Essential Oil Extracted from <em>Artemisia absinthium </em>L. (<em>Ariti</em>). <em>Clinical Biotechnology and Microbiology</em> 4 (1): 11–21.</p>
<p>Abegaz, B. & Yohannes, P.G. (1982) Constituents of the essential oil of <em>Artemisia rehan</em>. <em>Phytochemistry</em> 21: 1791–1793. https://doi.org/10.1016/S0031-9422(82)85065-6</p>
<p>Asfaw, N. & Demissew, S. (2015) Essential Oil Composition of Four <em>Artemisia </em>species from Ethiopia. <em>Bulletin of Chemical Society of Ethiop</em>ia 29 (1): 123–128. https://doi.org/10.4314/bcse.v29i1.11</p>
<p>Baldwin, B.G. & Markos, S. (1998) Phylogenetic utility of the External Transcribed Spacer (<em>ETS</em>) of 18S–26S rDNA: Congruence of <em>ETS</em> and <em>ITS</em> trees of <em>Calycadenia </em>(Compositae). <em>Molecular Phylogenetics and Evolution</em> 10: 449–463. https://doi.org/10.1006/mpev.1998.0545</p>
<p>Basta, A., Tzakou, O., Couladis, M. & Pavlovic, M. (2007) Chemical Composition of <em>A. absinthium</em> L<em>. </em>from Greece. <em>Journal of Essential Oil Research</em> 19: 316–318. https://doi.org/10.1080/10412905.2007.9699291</p>
<p>Bremer, K. & Humphries, C.J. (1993) Generic Monograph of the Asteraceae-Anthemideae. <em>Bulletin of Natural History Museum of London (Botany series)</em> 23: 71–177.</p>
<p>Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) “jModelTest 2: more models, new heuristics and parallel computing”. <em>Nature Methods</em> 9 (8): 772. https://doi.org/10.1038/nmeth.2109</p>
<p>Edgar, R.C. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput, <em>Nucleic Acids Research</em> 32 (5): 1792–1797. https://dio.org/10.1093/nar/gkh340</p>
<p>El-Moslimany, A.P. (1990) Ecological significance of common non arboreal pollen: examples from dry lands of the Middle East. <em>Review of Palaeobotany and Palynology</em> 64: 343–350. https://doi.org/10.1016/0034-6667(90)90150-H</p>
<p>Francisco-Ortega, J., Barber, J.C., Santos-Guerra, A., Febles-Hernandez, R. & Jansen, R.K. (2001) Origin and evolution of the endemic genera of Gonosperminae (<em>Asteracea</em>e: <em>Anthemideae</em>) from the Canary Islands: evidence from nucleotide sequences of the internal transcribed spacers of the nuclear ribosomal DNA. <em>American Journal of Botany</em> 88 (1): 161–169. https://doi.org/10.2307/2657136</p>
<p>Garcia, S., Garnatje, T., Hidalgo, O., McArthur, E.D., Siljak-Yakovlev, S. & Vallès, J. (2007) Extensive ribosomal DNA (18S5.8S-26S and 5S) co-localization in the North American endemic sagebrushes (subgenus <em>Tridentatae</em>, <em>Artemisia</em>, <em>Asteraceae</em>) revealed by FISH. <em>Plant Systematics and Evolution</em> 267: 79–92.</p>
<p>Garcia, S., Macarthur, E.D, Pellicer, J., Sanderson, S.C., Vallès, J. & Garnatje, T. (2011) A molecular phylogenetic approach to Western North America endemic <em>Artemisia</em> and allies (<em>Asteraceae</em>): Untangling the sagebrushes. <em>American Journal of Botany</em> 98: 638–653. https://dio.org.10.3732/ajb.1000386</p>
<p>Goloboff, P.A., Farris, J., Kallersjö, M., Oxelmann, B., Ramirez, M. & Suzumik, C. (2003) Improvements to resampling measures of group support. <em>Cladistics</em> 19: 324–332. https://dio.org/10.1111/j.1096-0031.tb00376.x</p>
<p>Goloboff, P.A., Farris, J.S. & Nixon, K.C. (2008) TNT a free program of phylogenetic analysis. <em>Cladistics</em> 24: 774–786. https://dio.org/10.1111/j.2008.00217.x</p>
<p>Guindon, S., Dufayared, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML3.0. <em>Systematic Biology</em> 59: 307–321.</p>
<p>Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95/98//NT. <em>Nucleic acids symposium series </em>41: 95–98.</p>
<p>Hayat, M.Q., Ashraf, M., Khan, M.A., Yasmin, G., Shaheen, N. & Jabeen, S. (2009a) Diversity of foliar trichomes and their systematic implications in the genus <em>Artemisia</em> (<em>Asteraceae</em>). <em>International Journal of Agriculture and Biology</em> 11: 542–546.</p>
<p>Hayat, M.Q., Khan, M.A., Ashraf, M. & Jabeen, S. (2009b) Ethnobotany of the genus <em>Artemisia</em> L. (<em>Asteraceae</em>) in Pakistan. <em>Ethnobotany Research and Applications</em> 7: 147–162.</p>
<p>Hobbs, C.R. & Baldwin, B.G. (2013) Asian origin and upslope migration of Hawaiian <em>Artemisia </em>(Compositae-Anthemideae). <em>Jornal of Biogeography</em> 40 (3): 442–454. https://doi.org./10.1016/j.jep.2010.05.062</p>
<p>Hussain, A., Potter, D., Kim, S., Muhammad, Q., Hayat, M.Q. & Bokhari, S.A.I. (2019) Molecular phylogeny of <em>Artemisia </em>(Asteraceae-Anthemideae) with emphasis on undescribed taxa from Gilgit-Baltistan (Pakistan) based on nrDNA (<em>ITS</em> and <em>ETS</em>) and cpDNA (<em>psbA-trnH</em>) sequences. <em>Plant Ecology and Evolution</em> 152 (3): 507–520. https://doi.org/10.5091/plecevo.2019.1583</p>
<p>Kornkven, A.B., Watson, L.E. & Estes, J.R. (1999) A molecular phylogeny of <em>Artemisia </em>section <em>Tridentatae </em>(Asteraceae) based on chloroplast DNA restriction site variation. <em>Systematic Botany </em>24 (1): 69–84. https://doi.org/10.2307/2419387</p>
<p>Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. <em>Molecular Biology and Evolution</em> 35: 1547–1549. https://doi.org/10.1093/molbev/msy096</p>
<p>Lachenmeier, D.W. (2010) Wormwood (<em>Artemisia absinthium</em> L.) A curious plant with both neurotoxic and neuroprotective properties? <em>Journal of Ethnopharmacology </em>131: 224–227. https://doi.org/10.1016/j.jep.2010.05.062</p>
<p>Ling, Y.R. (1994) The genera <em>Artemisia </em>L. and <em>Seriphidium </em>(Bess.) Poljak. in the world. <em>Compositae Newsletter</em> 25: 39–45.</p>
<p>Magdy, M.Y., Ahmed, A.M. & Yasair, S.S. (2015) Antimicrobial and Antioxidant activities of <em>Artemisia abyssinica</em> Extracts and DNA Degradation Effects. <em>Asian Journal of Biochemistry</em> 10 (1): 31–41. https://doi.org/10.3923/ajb.2015.31.41</p>
<p>Malik, S., Vitales, D., Hayat, M.Q., Korobkov, A.A., Garnatje, T. & Vallès, J. (2017) Phylogeny and biogeography of <em>Artemisia </em>subgenus <em>Seriphidium </em>(<em>Asteraceae; Anthemideae</em>). <em>Taxon </em>66 (4): 934–952. http://doi.org/10.12705/664.8</p>
<p>Markos, S. & Baldwin, B.G. (2001) Higher-level relationships and major lineages of <em>Lessingia</em> (<em>Compositae, Astereae</em>) based on nuclear rDNA internal and external transcribed spacer (<em>ITS</em> and <em>ETS</em>) sequences. <em>Systematic Botany</em> 26: 168–183.</p>
<p>Masuda, Y., Yukawa, T. & Kondo, K. (2009) Molecular phylogenetic analysis of members of Chrysanthemum and its related genera in the tribe Anthemideae, the Asteraceae in East Asia on the basis of the internal transcribed spacer (<em>ITS</em>) region and the external transcribed spacer (<em>ETS</em>) region of nrDNA. <em>Chromosome Botany</em> 4: 25–36. https://doi.org/10.3199/iscb.4.25</p>
<p>Mekonnen, M., Manahile, B. & Mengesha, B. (2015) “Screening of Botanical Extract for the Control of <em>Artemisia rehan</em> (<em>Artemisia absinthium</em>) Aphid (<em>Coloradoa absinthii Li</em>) in green house and field condition”. <em>Africa Journal of Crop Science </em>3 (7): 196–200.</p>
<p>Mohammadi, A., Sani, T., Ameri, A., Imani, M., Golmakani, E. & Kamali, H. (2015) Seasonal variation in the chemical composition, antioxidant activity and total phenolic content of <em>Artemisia absinthium </em>essential oils. <em>Pharmacognosy Research</em> 7 (4): 329–334. https://doi.org/10.4103/0974-8490.158441</p>
<p>Mukul, C. (2013) Chemical Investigations of the Essential Oils of Some <em>Artemisia</em> Species of Ethiopia. <em>IOSR Journal of Applied Chemistry </em>6 (4): 1–7. https://doi.org/10.9790/5736-0640107</p>
<p>Mulatu, A. & Mekonnen, Y. (2007) Spasmolytic effects of <em>Artemisia afra </em>and <em>Artemisia rehan </em>in tissue preparations. <em>Ethiopia Medical Journal </em>45 (4): 371–376.</p>
<p>Nguyen, H.T., Inotai, K., Radácsi, P., Tavaszi-Sárosi, S., Ladányi, M. & Zámboriné-Németh, É. (2017) Morphological, phytochemical and molecular characterization of intraspecific variability of wormwood (<em>Artemisia absinthium </em>L.). <em>Journal of Applied Botany and Food Quality</em> 90: 238–245. https://dio.org/10.5073/JABFQ.2017.090.030</p>
<p>Oberpreiler, C., Vogt, R. & Watson, L. (2003) Tribe <em>Anthemideae</em>.<em> In: </em>Kadereit, J. & Jeffrey, C. (eds.) <em>Families and Genera of Vascular plants</em>, vol. 8. Springer-Verlag, Berlin, pp. 342–374.</p>
<p>Pellicer, J., Garcia, S., Canela, M.A., Garnatje, T., Korobkov, A.A., Twibell, J.D. & Vallès, J. (2010) Genome size dynamics in <em>Artemisia</em> L. (Asteraceae): following the track of polyploidy. <em>Plant Biol</em>ogy 12 (5): 820–830. https://doi.org/10.1111/j.1438-8677.2009.00268.x</p>
<p>Pellicer, J., Garnatje, T. & Vallès, J. (2011) A<em>rtemisia </em>(<em>Asteraceae</em>): Understanding its evolution using cytogenetic and molecular systematic tools, with emphasis on subgenus <em>Dracunculus.</em> <em>Recent Advances in Pharmaceutical Sciences</em>: 199–222. [ISBN: 978-81-7895-528-5]</p>
<p>Pellicer, J., Hidalgo, O., Garnatje, T., Kondo, K. & Vallès, J. (2014) Life cycle versus systematic placement: Phylogenetic and cytogenetic studies in annual <em>Artemisia</em> (<em>Asteraceae</em>, <em>Anthemideae</em>). <em>Turkish Journal of Botany</em> 38: 1112–1122. https://doi.org/10.3906/bot-1404-102</p>
<p>Sanz, M., Schneeweiss, G., Vilatersana, R. & Vallès, J. (2011) Temporal origins and diversification of <em>Artemisia </em>and allies (<em>Anthemideae</em>, <em>Asteraceae</em>). <em>Collectanea Botanica (Barcelona) </em>30: 7–15. https://doi.org/10.3989/collectbot.2011.v30.001</p>
<p>Sanz, M., Vilatersana, R., Hidalgo, O., Garcia-Jacas, N., Susanna, A., Schneeweiss, G.M. & Vallès, J. (2008) Molecular phylogeny and evolution of floral characters of <em>Artemisia</em> and allies (<em>Anthemideae, Asteraceae</em>): evidence from nrDNA <em>ETS</em> and <em>ITS</em> sequence. <em>Taxon </em>57: 66–78.</p>
<p>Sarikhan, S., Aryakia, E., Azarbaijani, R., Dolatyari, A., Mousavi, S.H., Ramezani, H., Parsa Y., L., Shahzadeh, F.S.A., Naghavi, M.R. & Hosseini, S.G. (2012) Molecular phylogenetic analysis of the genus <em>Artemisia</em> L. (Asteraceae) in Iran based on nrDNA <em>ITS</em> and <em>ETS</em> sequences. Unpublished.</p>
<p>Tadesse, M. (2004) Asteraceae (Compositae). <em>In</em>: Hedberg, I., Friis, I. & Edwards, S. (Eds.) <em>Flora of Ethiopia and Eritrea</em>, vol. 4 (2). The National Herbarium, Addis Ababa University, Addis Ababa, Ethiopia & Uppsala, Sweden, pp. 221–224.</p>
<p>Tkach, N.V., Hoffmann, M.H., Röser, M. & Hagen, K.B. von (2008) Temporal patterns of evolution in the Arctic explored in <em>Artemisia</em> L. (<em>Asteraceae</em>) linage of different age. <em>Plant Ecology and Diversity </em>1: 161–169. https://doi.org/10.1080/17550802331912</p>
<p>Torrell, M., Garcia-Jacas, N., Susanna, A. & Vallès, J. (1999) Phylogeny in <em>Artemisia </em>L. (<em>Asteraceae, Anthemideae</em>) inferred from nuclear ribosomal (<em>ITS</em>) sequences. <em>Taxon </em>48 (4): 721–736. https://doi.org/10.2307/1223643</p>
<p>Vallès, J., Garcia, S., Hidalgo, O., Martín, J., Pellicer, J., Sanz, M. & Garnatje, T. (2011) Biology, genome evolution, biotechnological issues and research including applied perspectives in <em>Artemisia</em> (Asteraceae). <em>Advances Botany Research </em>60: 349–419. https://doi.org/10.1016/B978-0-12-385851-1.00015-9</p>
<p>Vallès, J. & McArthur, E.D. (2001) <em>Artemisia </em>systematics and phylogeny: cytogenetic and molecular in sights. <em>In</em>: McArthur, E.D. & Fairbanks, D.J. (Eds.) <em>Shrubland Ecosystem Genetics and Biodiversity</em>, Department of Agriculture, Forest Service, Rocky Mountain Research Station, pp. 67–74.</p>
<p>Vallès, J., Torrell, M., Garnatje, T., Garcia-Jacas, R., Vilatersana, N. & Susanna, A. (2003) Genus <em>Artemisia </em>and its allies, phylogeny of the subtribe <em>Artemisiinae</em> (<em>Asteraceae</em>, <em>Anthemiadeae</em>) based on nucleotide sequences of nuclear ribosomal DNA internal transcribed spacers (ITS). <em>Plant Biology</em> 5: 274–284. https://doi.org/10.1055/s-2003-40790</p>
<p>Wang, W.M. (2004) On the origin and development of <em>Artemisia </em>(<em>Asteraceae</em>) in the geological past. <em>Botanical Journal of the Linnean Society</em> 145: 331–336. https://doi.org/10.1111/j.1095-8339.2004.00287.x</p>
<p>Watson, L.E., Bates, P.L., Evans, T.M., Unwin, M.M. & Estes, J.R. (2002) Molecular phylogeny of subtribe <em>Artemisiinae</em> (<em>Asteraceae</em>) including <em>Artemisia </em>and its allied and segregate genera. <em>BMC Evolutionary Biology </em>2: e17. https://doi.org/10.1186/1471-2148-2-17</p>
<p>White, T.J., Bruns, T.D, Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>In</em>: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (eds.) <em>PCR protocols: a guide to methods and applications</em>. Academic Press, San Diego, California, U.S.A, pp. 315–322. https://doi.org/1016/B978-0-12-372180-8.50042-1</p>