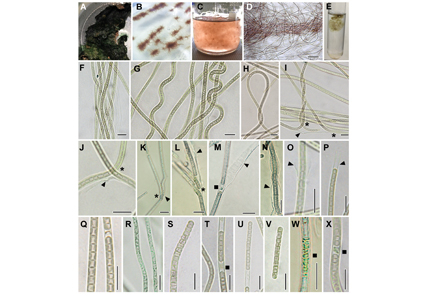
Abstract
Cyanobacterial diversity has been extensively studied; however, scant knowledge exists concerning genetic data and higher accuracy for tropical cyanobacterial diversity. In particular, the existence of unnamed monophyletic genera within the Oculatellaceae requires further investigation. Here, two novel strains (NUACC13 and NUACC14) were isolated from a cyanobacterial mat covering border soil of a small freshwater pond in Phitsanulok Province, Thailand. A polyphasic approach combining genetic (16S rDNA and rpoC1 gene phylogenies, evolutionary distance and ITS secondary structures) and morphological information was applied to evaluate the taxonomic status of these novel strains. All phylogenetic analyses of 16S rDNA and rpoC1 gene sequences placed our strains within the Oculatellaceae, strongly supporting a novel monophyletic clade that did not belong to other known Oculatellaceae genera. The 16S rDNA sequence similarity (<96.2%) values among the closest described genera also supported the novelty of our strains. The hypothetical secondary structures (D1–D1′, Box-B, V2 and V3 helices) of the ITS region confirmed the uniqueness of our strains, indicating differences from the related genera. Morphological features of the novel strains differed from the phylogenetically closest taxa by size and color of filament and vegetative cell and the presence of a frayed sheath. Considering all the results, we herein described our strains as the novel genus Siamcapillus gen. nov. in the Oculatellaceae with the type species S. rubidus sp. nov.
References
<p>Basheva, D., Moten, D., Stoyanov, P., Belkinova, D., Mladenov, R. & Teneva, I. (2018) Content of phycoerythrin, phycocyanin, alophycocyanin and phycoerythrocyanin in some cyanobacterial strains: Applications. <em>Engineering in Life Sciences</em> 18: 861–866. https://doi.org/10.1002/elsc.201800035</p>
<p>Bennett, A. & Bogorad, L. (1973) Complementary chromatic adaptation in a filamentous blue-green alga. <em>Journal of Cell Biology</em> 58: 419–435. https://doi.org/10.1083/jcb.58.2.419</p>
<p>Berthold, D.E., Lefler, F.W. & Laughinghouse, H.D. (2021) Untangling filamentous marine cyanobacterial diversity from the coast of South Florida with the description of <em>Vermifilaceae</em> fam. nov. and three new genera: <em>Leptochromothrix</em> gen. nov., <em>Ophiophycus</em> gen. nov., and <em>Vermifilum</em> gen. nov. <em>Molecular Phylogenetics and Evolution</em> 160: 107010. https://doi.org/10.1016/j.ympev.2020.107010</p>
<p>Berrendero Gómez, E., Johansen, J.R., Kaštovský, J., Bohunická, M. and Čapková, K. (2016) <em>Macrochaete</em> gen. nov. (Nostocales, Cyanobacteria), a taxon morphologically and molecularly distinct from <em>Calothrix</em>. <em>Journal of Phycology</em> 52: 638–655. https://doi.org/10.1111/jpy.12425</p>
<p>Brito, Â., Ramos, V., Mota, R., Lima, S., Santos, A., Vieira, J., Vieira, C.P., Kaštovský, J., Vasconcelos, V.M. & Tamagnini, P. (2017) Description of new genera and species of marine cyanobacteria from the Portuguese Atlantic coast, <em>Molecular Phylogenetics and Evolution</em> 111: 18–34. https://doi.org/10.1016/j.ympev.2017.03.006.</p>
<p>Brown, I.I., Bryant, D.A., Casamatta, D., Thomas-Keprta, K.L., Sarkisova, S.A., Shen, G., Graham, J.E., Boyd, E.S., Peters, J.W., Garrison, D.H. & McKay, D.S. (2010) Polyphasic characterization of a thermotolerant siderophilic filamentous cyanobacterium that produces intracellular iron deposits. <em>Applied Environmental Microbiology</em> 76: 6664–6672. https://doi.org/10.1128/AEM.00662-10</p>
<p>Casamatta, D., Johansen, J.R., Vis, M.L. & Broadwater, S.T. (2005) Molecular and morphological characterization of ten polar and near polar strains within the Oscillatoriales (Cyanobacteria). <em>Journal of Phycology</em> 41: 421–438. https://doi.org/10.1111/j.1529-8817.2005.04062.x</p>
<p>Chakraborty, S., Maruthanayagam, V., Achari, A., Pramanik, A., Jaisankar, P. & Mukherjee, J. (2021) <em>Aerofilum fasciculatum</em> gen. nov., sp. nov. (Oculatellaceae) and <em>Euryhalinema pallustris</em> sp. nov. (Prochlorotrichaceae) isolated from an Indian mangrove forest. <em>Phytotaxa</em> 522: 165–186. https://doi.org/10.11646/phytotaxa.522.3.1</p>
<p>Chan, P.P., & Lowe, T.M. (2019) tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences<em>. Methods in molecular biology</em> <em>(Clifton, N.J.)</em> 1962: 1–14. https://doi.org/10.1007/978-1-4939-9173-0_1</p>
<p>Charpy, L., Casareto, B.E., Langlade, M.J. & Suzuki, Y. (2012) Cyanobacteria in coral reef ecosystems: A review. <em>Journal of Marine Biology</em> 2012: 1–9. https://doi.org/10.1155/2012/259571</p>
<p>Chatchawan, T., Komárek, J., Strunecky, O., Smarda, J. & Peerapornpisal, Y. (2012) <em>Oxynema</em>, a new genus separated from the genus <em>Phormidium</em> (Cyanophyta). <em>Cryptogamie Algologie</em> 33: 41–59. https://doi.org/10.7872/crya.v33.iss1.2011.041</p>
<p>Demay, J., Bernard, C. Reinhardt, A. & Marie, B. (2019) Natural products from cyanobaceria: Focus on beneficial activities. <em>Marine Drugs</em> 17: 320. https://doi.org/10.3390/md17060320</p>
<p>Drancourt, M., Bollet, C., Carlioz, A., Martelin, R., Gayral, J.P. & Raoult, D. (2000) 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial Isolates. <em>Journal of Clinical Microbiology</em> 38: 3623–3630. https://doi.org/10.1128/JCM.38.10.3623-3630.2000</p>
<p>Dvořák, P., Poulíčková, A., Hašler, P., Belli, M., Casamatta, D.A. & Papini, A. (2015) Species concepts and speciation factors in cyanobacteria, with connection to the problems of diversity and classification. <em>Biodiversity and Conservation</em> 24: 739–757. https://doi.org/10.1007/s10531-015-0888-6</p>
<p>Dvořák, P., Hašler, P., Casamatta, D.A. & Poulíčková, A. (2021) Underestimated cyanobacteria diversity: trends and perspectives of research in tropical environments. <em>Fottea</em> 21: 110–127. https://doi.org/10.0.21.131/fot.2021.009</p>
<p>Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. <em>Nucleic Acids Research</em> 32: 1792–1797. https://doi.org/10.1093/nar/gkh340</p>
<p>Genuário, D.B., Vas, M.G.M.V., Hentschke, G.S., Sant’Anna, C.L. & Fiore, M.F. (2015) <em>Halotia</em> gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. <em>International Journal of Systematic and Evolutionary Microbiology</em> 65: 663–675. https://doi.org/10.1099/ijs.0.070078-0</p>
<p>González-Resendiz, L., Johansen, J.R., Alba-Lois, L., Segal-Kischinevzky, C., Escobar-Sámche, V., Jiménez-García, L.F., Huaer, T. & León-Tejera, H. (2018) <em>Nunduva</em>, a new marine genus of Rivulariaceae (Nostocales, Cyanobacteria) from marine rocky shores. <em>Fottea</em> 18: 86–105. https://doi.org/10.5507/fot.2017.018</p>
<p>González-Resendiz, L., Johansen, J.R., León-Tejera, H., Sánchez, L., Segal-Kischinevzky, C., Escobar-Sánchez, V. & Morales, M. (2019) A bridge too far in naming species: a total evidence approach does not support recognition of four species in <em>Desertifilum</em> (Cyanobacteria). <em>Journal of Phycology</em> 55: 898–911. https://doi.org/10.1111/jpy.12867</p>
<p>Guindon, S., Dufayard, J-F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. <em>Systematic Biology</em> 59: 307–321. https://doi.org/10.1093/sysbio/syq010</p>
<p>Hašler, P., Dvořák, P., Johansen, J.R., Kitner, M., Ondřej, V. & Poulíčková, A. (2012) Morphological and molecular study of epipelic filamentous genera <em>Phormidium</em>, <em>Microcoleus</em> and <em>Geitlerinema</em> (Oscillatoriales, Cyanophyta/Cyanobacteria). <em>Fottea</em> 12: 341–356. https://doi.org/10.5507/fot.2012.024</p>
<p>Hašler, P., Dvořák, P., Poulíčková, P. & Casamatta, D.A. (2014) A novel genus <em>Ammassolinea </em>gen. nov. (Cyanobacteria) isolated from sub-tropical epipelic habitats. <em>Fottea</em> 14: 241–248. https://doi.org/10.5507/fot.2014.018</p>
<p>Hauerová, R., Hauer, T., Kaštovský, J., Komárek, J., Lepšová-Skácelová, O. & Mareš, O. (2021) <em>Tenebriella</em> gen. nov. – The dark twin of <em>Oscillatoria</em>, <em>Molecular Phylogenetics and Evolution</em> 165: 107293. https://doi.org/10.1016/j.ympev.2021.107293.</p>
<p>Iteman, I., Rippka, R., de Marsac, N.T. & Herdman, M. (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. <em>Microbiology</em> 146: 1275–1286. https://doi.org/10.1099/00221287-146-6-1275</p>
<p>Jahodářová, E., Dvořák, P., Hašler, P., Holušová, K. & Poulíčková, A. (2018) <em>Elainella</em> gen. nov.: a new tropical cyanobacterium characterized using a complex genomic approach. <em>European Journal of Phycology</em> 53: 39–51 https://doi.org/10.1080/09670262.2017.1362591</p>
<p>Johansen, J.R. & Casamatta, D.A. (2005) Recognizing cyanobacterial diversity through adoption of a new species paradigm. <em>Algological Studies</em> 117: 71–93. https://doi.org/10.1127/1864-1318/2005/0117-0071</p>
<p>Johansen, J.R, Kovácik, L., Casamatta, D.A., Fuĉiková, K. & Kaštovský, J. (2011) Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: <em>Leptolyngbya corticola</em> sp. nov. (Pseudanabaenaceae, Cyanobacteria). <em>Nova Hedwigia</em> 92: 283–302. https://doi.org/10.1127/0029-5035/2011/0092-0283</p>
<p>Johansen, J.R., González-Resendiz, L., Escobar-Sánchez, V., Segal-Kischinevzky, C., Martínez-Yerena, J., Hernández-Sánchez, J., Hernández-Pérez, G. & León-Tejera, H. (2021) When will taxonomic saturation be achieved? A case study in <em>Nunduva</em> and <em>Kyrtuthrix </em>(Rivulariaceae, Cyanobacteria). <em>Journal of Phycology</em> 57 (6): 1699–1720. https://doi.org/10.1111/jpy.13201</p>
<p>Jungblut, A., Lovejoy, C. & Vincent, W. (2010) Global distribution of cyanobacterial ecotypes in the cold biosphere. <em>The ISME Journal</em> 4: 191–202. https://doi.org/10.1038/ismej.2009.113</p>
<p>Komárek, J. (2006) Cyanobacterial Taxonomy: Current problems and prospects for the integration of traditional and molecular approaches. <em>Algae</em> 21: 349–375. https://doi.org/10.4490/algae.2006.21.4.349</p>
<p>Komárek, J. & Anagnostidis, K. (2005) Cyanoprokaryota.2.Oscillatoriales.<em> In: </em>Büdel, B., Krienitz, L., Gärtner, G. & Schagerl, M. (Eds.) Süsswasserflora von Mitteleuropa 19/2. Elsevier/Spektrum, Heidelberg. pp. 759.</p>
<p>Kaštovský, J., Berrendero Gomez, E., Hladil, J. & Johansen, J.R. (2014) <em>Cyanocohniella calida</em> gen. et sp. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. <em>Phytotaxa</em> 181: 279–292. https://doi.org/10.11646/phytotaxa.181.5.3</p>
<p>Komárek, J., Kaštovsky´, J., Mareš, J. & Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. <em>Preslia</em> 86: 295–335. [http://www.preslia.cz/P144Komarek.pdf]</p>
<p>Komárek, J. (2016) A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. <em>European Journal of Phycology</em> 51: 346–353. https://doi.org/10.1080/09670262.2016.1163738</p>
<p>Komárek, J. (2018) Several problems of the polyphasic approach in the modern cyanobacterial system. <em>Hydrobiologia</em> 811: 7–17. https://doi.org/10.1007/s10750-017-3379-9</p>
<p>Kozlíková-Zapomělová, E., Chatchawan, T., Kaštovský, J., & Komárek, J. (2016) Phylogenetic and taxonomic position of the genus <em>Wollea</em> with the description of <em>Wollea salina</em> sp. nov. (Cyanobacteria, Nostocales). <em>Fottea</em> 16: 43–55. https://doi.org/10.5507/fot.2015.026</p>
<p>Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. <em>Molecular Biology and Evolution</em> 35: 1547–1549. https://doi.org/10.1093/molbev/msy096</p>
<p>Lyons, T.W., Reinhard, C.T. & Planavsky, N.J. (2014) The rise of oxygen in Earth’s early ocean and atmosphere. <em>Nature</em> 506: 307–315. https://doi.org/10.1038/nature13068</p>
<p>Mai, T., Johansen, J.R., Pietrasiak, N., Bohunicka, M. & Martin, M.P. (2018) Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae <em>fam. nov.</em> and Trichocoleaceae <em>fam. nov.</em> and six new genera containing 14 species. <em>Phytotaxa</em> 365: 1–59. https://doi.org/10.11646/phytotaxa.365.1.1</p>
<p>Malone, C.F.d.S., Genuário, D.B., Vaz, M.G.M.V., Fiore, M.F. & Sant’Anna, C.L. (2021) <em>Monilinema</em> gen. nov., a homocytous genus (Cyanobacteria, Leptolyngbyaceae) from saline–alkaline lakes of Pantanal wetlands, Brazil. <em>Journal of Phycology</em> 57: 473–483. https://doi.org/10.1111/jpy.13106</p>
<p>Mareš, J., Johansen, J.R., Hauer, T., Zima Jr., J., Ventura, S., Cuzman, O., Tiribilli, B & Kaštovský, J. (2019) Taxonomic resolution of the genus <em>Cyanothece</em> (Chroococcales, Cyanobacteria), with a treatment on <em>Gloeothece </em>and three new genera, <em>Crocosphaera</em>, <em>Rippkaea</em>, and <em>Zehria</em>. <em>Journal of Phycology</em> 55: 578−610. https://doi.org/10.1111/jpy.12853</p>
<p>Mathews Lab, University of Rochester Medical Center (2018) RNA structure, version 6.0.1 Available from: https://rna.urmc.rochester.edu/RNAstructure.html (accessed 16 April 2018)</p>
<p>Mühlsteinová, R., Johansen, J.R., Pietrasiak, N., Martin, M.P., Osoirio-Santos, K. & Warren, S.D. (2014) Polyphasic characterization of <em>Trichocoleus desertorum</em> sp. nov. Pseudanabaenales, Cyanobacteria) from desert soils and phylogenetic placement of the genus <em>Trichocoleus</em>. <em>Phytotaxa </em>163: 241–261. https://doi.org/10.11646/phytotaxa.163.5.1</p>
<p>Neilan, B.A., Jacobs, D., Dot, T.D., Blackall, L.L., Hawkins, P.R., Cox, P.T. & Goodman, A.E. (1997) rRNA Sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus <em>Microcystis</em>. <em>International Journal of Systematic Bacteriology</em> 47: 693–697. https://doi.org/10.1099/00207713-47-3-693</p>
<p>Nowruzi, B., & Shalygin, S. (2021) Multiple phylogenies reveal a true taxonomic position of <em>Dulcicalothrix alborzica</em> sp. nov. (Nostocales, Cyanobacteria). <em>Fottea</em> 21: 235–246. https://doi.org/10.5507/fot.2021.008</p>
<p>Nübel, U., Garcia-Pichel, F. & Muyzer, G. (1997) PCR primers to amplify 16S rRNA genes from cyanobacteria. <em>Applied and Environmental Microbiology</em> 63: 3327–3332. https://doi.org/10.1128/aem.63.8.3327-3332.1997</p>
<p>Nylander, J.A.A. (2008) mrmodeltest v2. Program distributed by author. Evolutionary Biology Centre, Uppsala University.</p>
<p>Osorio-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kovacik, L., Martin, M.P. & Johansen, J.R. (2014) Seven new species of <em>Oculatella</em> (Pseudanabaenales, Cyanobacteria) <em>European Journal of Phycology</em> 49: 450–470. https://doi.org/10.1080/09670262.2014.976843</p>
<p>Pagel, F., Guedes, A.C., Amaro, H.M., Kijjoa, A. & Vasconcelos, V. (2019) Phycobiliproteins from cyanobacteria: Chemistry and biotechnological applications. <em>Biotechnology Advances</em> 37: 422–443. https://doi.org/10.1016/j.biotechadv.2019.02.010.</p>
<p>Partensky, F., Six, C., Ratin, M., Garczarek, L., Vaulot, D., Probert, I., Calteau, A., Gourvil, P., Marie, D., Grébert, T., Bouchier, C., Le Panse, S., Gachenot, M., Rodríguez, F. & Garrido, J.L. (2018) A novel species of the marine cyanobacterium <em>Acaryochloris</em> with a unique pigment content and lifestyle. <em>Scientific Report</em> 8: 9142. https://doi.org/10.1038/s41598-018-27542-7</p>
<p>Pietrasiak, N., Muhlsteinova, R., Siegesmund, M.A. & Johansen, J.R. (2014) Phylogenetic placement of <em>Symplocastrum </em>(Phormidiaceae) with a new combination <em>S. californicum </em>and two new species: <em>S. flechtnerae </em>and <em>S. torsivum</em>. <em>Phycologia </em>53: 529–541. https://doi.org/10.2216/14-029.1</p>
<p>Pietrasiak, N., Osorio-Santos, K., Shalygin, S., Martin, M.P. & Johansen, J.R. (2019) When is a lineage a species? A case study in <em>Myxacorys</em> gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. <em>Journal of Phycology</em> 55: 976–996. https://doi.org/10.1111/jpy.12897</p>
<p>Pietrasiak, N., Reeve, S., Osorio-Santos, K., Lipson, D.A. & Johansen, J.R. (2021) <em>Trichotorquatus</em> gen. nov. - a new genus of soil cyanobacteria discovered from American drylands. <em>Journal of Phycology</em> 57: 886–902. https://doi.org/10.1111/jpy.13147</p>
<p>Philippot, L., Andersson, S.G., Battin, T.J., Prosser, J.I., Schimel, J.P., Whitman, W.B. & Hallin, S. (2010) The ecological coherence of high bacterial taxonomic ranks. <em>Nature Reviews Microbiology</em> 8: 523–529. https://doi.org/10.1038/nrmicro2367</p>
<p>Sciuto, K. & Moro, I. (2016) Detection of the new cosmopolitan genus <em>Thermoleptolyngbya</em> (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S–23S ITS region. <em>Molecular Phylogenetics and Evolution</em> 105: 15–35. https://doi.org/10.1016/j.ympev.2016.08.010</p>
<p>Sciuto, K., Moschin, E. & Moro, I. (2017) Cryptic Cyanobacterial Diversity in the Giant Cave (Trieste, Italy): The New Genus <em>Timaviella</em> (Leptolyngbyaceae). <em>Cryptogamie Algologie</em> 38: 285–323. https://doi.org/10.7872/crya/v38.iss4.2017.285</p>
<p>Sendall, B.C. & McGregor, G.B. (2018) Cryptic diversity within the <em>Scytonema</em> complex: Characterization of the paralytic shellfish toxin producer <em>Heterosyctonema crispum</em>, and the establishment of the family Heteroscytonemataceae (Cyanobacteria/Nostocales). <em>Harmful Algae</em> 80: 158–170. https://doi.org/10.1016/j.hal.2018.11.002.</p>
<p>Seo, P-S. & Yokota, A. (2003) The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, <em>gyrB, rpoC1</em> and <em>rpoD1</em> gene sequences. <em>The journal of General and Applied Microbiology </em>49: 191–203. https://doi.org/10.2323/jgam.49.191</p>
<p>Shen, L.-Q., Zhang, Z.-C., Shang, J.-L., Li, Z.-K., Chen, M., Li, R. & Qiu, B.-S. (2022) <em>Kovacikia minuta</em> sp. nov. (Leptolyngbyaceae, Cyanobacteria), a new freshwater chlorophyll f-producing cyanobacterium. <em>Journal of Phycology</em> 58 (3): 424–435. https://doi.org/10.1111/jpy.13248</p>
<p>Strunecky, O., Bohunická, M., Johansen, J.R., Čapková, K., Raabová, L., Dvořák, P., & Komárek, J. (2017) A revision of the genus <em>Geitlerinema</em> and a description of the genus <em>Anagnostidinema</em> gen. nov. (Oscillatoriophycidae, Cyanobacteria). <em>Fottea</em> 17: 114–126. https://doi.org/10.5507/fot.2016.025</p>
<p>Strunecky, O., Raabova, L., Bernardova, A., Ivanova, A.P., Semanova, A., Crossley, J. & Kaftan, D. (2020) Diversity of cyanobacteria at the Alaska North Slope with description of two new genera: <em>Gibliniella</em> and <em>Shackletoniella</em>, <em>FEMS Microbiology Ecology </em>96: fiz189. https://doi.org/10.1093/femsec/fiz189</p>
<p>Strunecky, O., Kopejtka, K., Goecke, F., Tomasch, J., Lukavský, J., Neori, A., Kahl, S., Pieper, D.H., Pilarski, P., Kaftan, D. & Koblížek, M. (2019) High diversity of thermophilic cyanobacteria in Rupite hot spring identified by microscopy, cultivation, single-cell PCR and amplicon sequencing. <em>Extremophiles </em>23: 35–48. https://doi.org/10.1007/s00792-018-1058-z.</p>
<p>Ronquist, F., Teslenko, M. van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Tang, W., Cerdán-García, E., Berthelot, H., Polyviou, D., Wang, S., Baylay, A., Whitby, H., Planquette, H., Mowlem, M., Robidart, J., & Cassar, N. (2020) New insights into the distributions of nitrogen fixation and diazotrophs revealed by high-resolution sensing and sampling methods. <em>The ISME journal</em> 14: 2514–2526. https://doi.org/10.1038/s41396-020-0703-6</p>
<p>Tawong, W. (2017) Diversity of the Potential 2-Methylisoborneol-Producing Genotypes in Thai Strains of <em>Planktothricoides</em> (Cyanobacteria). <em>Brazilian Archives of Biology and Technology</em> 60: e17160567. https://doi.org/10.1590/1678-4324-2017160567</p>
<p>Tawong, W., Pongcharoen, P., Pongpadung, P. & Ponza, S. (2019a) <em>Neowollea manoromense</em> sp. nov. (Nostocales, Cyanobacteria), a novel geosmin producer isolated from Thailand. <em>Phytotaxa</em> 424: 1–17.</p>
<p>Tawong, W., Pongcharoen, P., Nishimura, T. & Adachi, M. (2019b) Molecular characterizations of Thai <em>Raphidiopsis raciborskii</em> (Nostocales, Cyanobacteria) based on 16S rDNA, <em>rbcLX</em>, and cylindrospermopsin synthetase genes. <em>Plankton and Benthos Research</em> 14: 211–223. https://doi.org/10.3800/pbr.14.211</p>
<p>Wang, Y., Cai, F., Jia, N. & Li, R. (2019) Description of a novel coccoid cyanobacterial genus and species <em>Sinocapsa zengkensis</em> gen. nov. sp. nov. (Sinocapsaceae, incertae sedis), with taxonomic notes on genera in Chroococcidiopsidales. <em>Phytotaxa </em>409: 146–160. https://doi.org/10.11646/phytotaxa.409.3.3</p>
<p>Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.-H., Whitman, W.B., Euzéby, J., Rudolf, A. & Rosselló-Móra, R. (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. <em>Nature Reviews Microbiology</em> 12: 635–645. https://doi.org/10.1038/nrmicro3330</p>
<p>Zammit, G., Billi, D. & Albertano, P. (2012) The subaerophytic cyanobacterium <em>Oculatella subterranea</em> (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: a cytomorphological and molecular description. <em>European Journal of Phycology</em> 47: 341–354. https://doi.org/10.1080/09670262.2012.717106</p>
<p>Zammit, G. (2018) Systematics and biogeography of sciophilous cyanobacteria; an ecological and molecular description of <em>Albertania skiophila</em> (Leptolyngbyaceae) gen. & sp. nov. <em>Phycologia</em> 57: 481–491. https://doi.org/10.2216/17-125.1</p>