Abstract
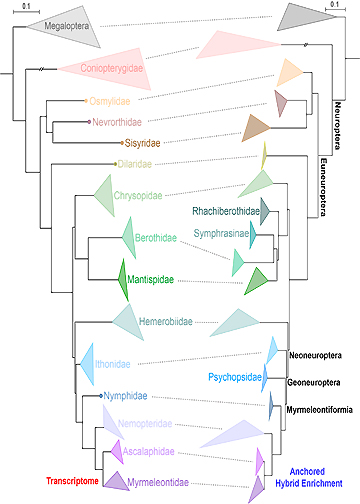
Exponential growth of large-scale data for Neuropterida, an iconic group of insects used in behavioural, ecological, and evolutionary studies, has greatly changed our understanding of the origin and evolution of lacewings and their allies. Recent phylogenomic studies of Neuropterida based on mitogenomes, anchored hybrid enrichment (AHE) data, and transcriptomes have yielded a well-resolved and largely congruent phylogeny. Some interfamilial relationships of lacewings, however, remain inconsistent among different phylogenomic studies. Here we re-analysed the genome-scale AHE and transcriptomic data for Neuropterida under the better fitting site-heterogeneous CAT-GTR+G model and recovered a strongly supported and congruent tree for the deeper phylogeny of Neuroptera. Integrating the smaller but more broadly sampled AHE and the larger but less-sampled transcriptomic data, we present a holistic phylogeny of Neuropterida from which to explore patterns of evolution across the clade. Our re-analyses of the largest available datasets of Neuropterida highlight the significance of modelling across-site compositional heterogeneity and model comparison in large-scale phylogenomic studies of insects.
References
- Aspöck, U., Haring, E. & Aspöck, H. (2012) The phylogeny of the Neuropterida: long lasting and current controversies and challenges (Insecta: Endopterygota). Arthropod Systematics & Phylogeny, 70, 119–129.
- Badano, D., Aspöck, U., Aspöck, H. & Cerretti, P. (2017) Phylogeny of Myrmeleontiformia based on larval morphology (Neuropterida: Neuroptera): Phylogeny of Myrmeleonti-formia. Systematic Entomology, 42, 94–117. https://doi.org/10.1111/syen.12200
- Badano, D., Engel, M.S., Basso, A., Wang, B. & Cerretti, P. (2018) Diverse Cretaceous larvae reveal the evolutionary and behavioural history of antlions and lacewings. Nature Communications, 9, 3257. https://doi.org/10.1038/s41467-018-05484-y
- Cai, C.Y., Tihelka, E., Giacomelli, M., Lawrence, J.F., Ślipiński, A., Kundrata, R., Yamamoto, S., Thayer, M.K., Newton, A.F., Leschen, R.A.B., Gimmel, M.L., Lü, L., Engel, M.S., Bouchard, P., Huang, D.Y., Pisani, D. & Donoghue, P.C.J. (2022) Integrated phylogenomics and fossil data illuminate the evolution of beetles. Royal Society Open Science, 9, 211771. https://doi.org/10.1098/rsos.211771
- Cai, C.Y., Tihelka, E., Pisani, D. & Donoghue, P.C.J. (2020) Data curation and modeling of compositional heterogeneity in insect phylogenomics: A case study of the phylogeny of Dytiscoidea (Coleoptera: Adephaga). Molecular Phylogenetics and Evolution, 147, 106782. https://doi.org/10.1016/j.ympev.2020.106782
- Criscuolo, A. & Gribaldo, S. (2010) BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology, 10, 210. https://doi.org/10.1186/1471-2148-10-210
- Engel, M.S., Winterton, S.L. & Breitkreuz, L.C.V. (2018) Phylogeny and evolution of Neuropterida: where have wings of lace taken us? Annual Review of Entomology, 63, 531–551. https://doi.org/10.1146/annurev-ento-020117-043127
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution, 35, 518–522. https://doi.org/10.1093/molbev/msx281
- Inagaki, Y. & Roger, A.J. (2006) Phylogenetic estimation under codon models can be biased by codon usage heterogeneity. Molecular Phylogenetics and Evolution, 40, 428–434. https://doi.org/10.1016/j.ympev.2006.03.020
- Jones, J.R. (2019) Total-evidence phylogeny of the owlflies (Neuroptera, Ascalaphidae) supports a new higher-level classification. Zoologica Scripta, 48, 761–782. https://doi.org/10.1111/zsc.12382
- Kapli, P., Yang, Z.H. & Telford, M.J. (2020) Phylogenetic tree building in the genomic age. Nature Reviews Genetics, 21, 428–444. https://doi.org/10.1038/s41576-020-0233-0
- Lartillot, N. (2020) PhyloBayes: Bayesian phylogenetics using site-heterogeneous models. In: Scornavacca, C., Delsuc, F. & Galtier, N. (Eds), Phylogenetics in the Genomic Era. No commercial publisher, Authors open access book, pp.1.5:1–1.5:16. Available at: https://hal.archives-ouvertes.fr/hal-02535342 (Accessed 10 Feb 2023)
- Lartillot, N. (2022) Identifying the best approximating model in Bayesian phylogenetics: Bayes factors, cross-validation or wAIC? bioRxiv, 2022.04.22.489153. https://doi.org/10.1101/2022.04.22.489153
- Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. (2013) PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Systematic Biology, 62, 611–615. https://doi.org/10.1093/sysbio/syt022
- Le, S.Q., Dang, C.C. & Gascuel, O. (2012) Modeling protein evolution with several amino acid replacement matrices depending on site rates. Molecular Biology and Evolution, 29, 2921–2936. https://doi.org/10.1093/molbev/mss112
- Lozano-Fernandez, J. (2022) A practical guide to design and assess a phylogenomic study. Genome Biology and Evolution, 14 (9), evac129. https://doi.org/10.1093/gbe/evac129
- Machado, R.J.P., Gillung, J.P., Winterton, S.L., Garzón-Orduña, I.J., Lemmon, A.R., Lemmon, E.M. & Oswald, J.D. (2019) Owlflies are derived antlions: anchored phylogenomics supports a new phylogeny and classification of Myrmeleontidae (Neuroptera). Systematic Entomology, 44, 418–450. https://doi.org/10.1111/syen.12334
- McKenna, D.D., Shin, S., Ahrens, D., Balke, M., Beza-Beza, C., Clarke, D.J., Donath, A., Escalona, H.E., Friedrich, F., Letsch, H., Liu, S.L., Maddison, D., Mayer, C., Misof, B., Murin, P.J., Niehuis, O., Peters, R.S., Podsiadlowski, L., Pohl, H., Scully, E.D., Yan, E.V., Zhou, X., Ślipiński, A. & Beutel, R.G. (2019) The evolution and genomic basis of beetle diversity. Proceedings of the National Academy of Sciences, 116, 24729–24737. https://doi.org/10.1073/pnas.1909655116
- Meusemann, K., Trautwein, M., Friedrich, F., Beutel, R.G., Wiegmann, B.M., Donath, A., Podsiadlowski, L., Petersen, M., Niehuis, O., Mayer, C., Bayless, K.M., Shin, S., Liu, S.L., Hlinka, O., Minh, B.Q., Kozlov, A., Morel, B., Peters, R.S., Bartel, D., Grove, S., Zhou, X., Misof, B. & Yeates, D.K. (2020) Are fleas highly modified Mecoptera? Phylogenomic resolution of Antliophora (Insecta: Holometabola). bioRxiv, 2020.11.19.390666. https://doi.org/10.1101/2020.11.19.390666
- Misof, B., Liu, S.L., Meusemann, K., Peters, R.S., Donath, A., Mayer, C., Frandsen, P.B., Ware, J., Flouri, T., Beutel, R.G., Niehuis, O., Petersen, M., Izquierdo-Carrasco, F., Wappler, T., Rust, J., Aberer, A.J., Aspöck, U., Aspöck, H., Bartel, D., Blanke, A., Berger, S., Böhm, A., Buckley, T.R., Calcott, B., Chen, J.Q., Friedrich, F., Fukui, M., Fujita, M., Greve, C., Grobe, P., Gu, S.C., Huang, Y., Jermiin, L.S., Kawahara, A.Y., Krogmann, L., Kubiak, M., Lanfear, R., Letsch, H., Li, Y.Y., Li, Z.Y., Li, J.G., Lu, H.R., Machida, R., Mashimo, Y., Kapli, P., McKenna, D.D., Meng, G.L., Nakagaki, Y., Navarrete-Heredia, J.L., Ott, M., Ou, Y.X., Pass, G., Podsiadlowski, L., Pohl, H., von Reumont, B.M., Schütte, K., Sekiya, K., Shimizu, S., Slipinski, A., Stamatakis, A., Song, W.H., Su, X., Szucsich, N.U., Tan, M.H., Tan, X.M., Tang, M., Tang, J.B., Timelthaler, G., Tomizuka, S., Trautwein, M., Tong, X.L., Uchifune, T., Walzl, M.G., Wiegmann, B.M., Wilbrandt, J., Wipfler, B., Wong, T.K.F., Wu, Q., Wu, G.X., Xie, Y.L., Yang, S.Z,, Yang, Q., Yeates, D.K., Yoshizawa, K., Zhang, Q., Zhang, R., Zhang, W.W., Zhang, Y.H., Zhao, J., Zhou, C.R., Zhou, L.L., Ziesmann, T., Zou, S.J., Li, Y.R., Xu, X., Zhang, Y., Yang, H.M., Wang, J., Wang, J., Kjer, K.M. & Zhou, X. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science, 346, 763–767. https://doi.org/10.1126/science.1257570
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
- Philippe, H., Brinkmann, H., Lavrov, D.V., Littlewood, D.T.J., Manuel, M., Wörheide, G. & Baurain, D. (2011) Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biology, 9, e1000602. https://doi.org/10.1371/journal.pbio.1000602
- Rota-Stabelli, O., Lartillot, N., Philippe, H. & Pisani, D. (2013) Serine codon-usage bias in deep phylogenomics: Pancrustacean relationships as a case study. Systematic Biology, 62, 121–133. https://doi.org/10.1093/sysbio/sys077
- Schwentner, M., Combosch, D.J., Pakes Nelson, J. & Giribet, G. (2017) A phylogenomic solution to the origin of insects by resolving crustacean-hexapod relationships. Current Biology, 27, 1818–1824. https://doi.org/10.1016/j.cub.2017.05.040
- Tihelka, E., Cai, C.Y., Giacomelli, M., Lozano-Fernandez, J., Rota-Stabelli, O., Huang, D.Y., Engel, M.S., Donoghue, P.C.J. & Pisani, D. (2021) The evolution of insect biodiversity. Current Biology, 31, R1299–R1311. https://doi.org/10.1016/j.cub.2021.08.057
- Tihelka, E., Giacomelli, M., Huang, D.Y., Pisani, D., Donoghue, P.C.J. & Cai, C.Y. (2020) Fleas are parasitic scorpionflies. Palaeoentomology, 3 (6), 641–653. https://doi.org/10.11646/palaeoentomology.3.6.16
- Vasilikopoulos, A., Balke, M., Beutel, R.G., Donath, A., Podsiadlowski, L., Pflug, J.M., Waterhouse, R.M., Meusemann, K., Peters, R.S., Escalona, H.E., Mayer, C., Liu, S.L., Hendrich, L., Alarie, Y., Bilton, D.T., Jia, F., Zhou, X., Maddison, D.R., Niehuis, O. & Misof, B. (2019) Phylogenomics of the superfamily Dytiscoidea (Coleoptera: Adephaga) with an evaluation of phylogenetic conflict and systematic error. Molecular Phylogenetics and Evolution, 135, 270–285. https://doi.org/10.1016/j.ympev.2019.02.022
- Vasilikopoulos, A., Misof, B., Meusemann, K., Lieberz, D., Flouri, T., Beutel, R.G., Niehuis, O., Wappler, T., Rust, J., Peters, R.S., Donath, A., Podsiadlowski, L., Mayer, C., Bartel, D., Böhm, A., Liu, S.L., Kapli, P., Greve, C., Jepson, J.E., Liu, X.Y., Zhou, X., Aspöck, H. & Aspöck, U. (2020) An integrative phylogenomic approach to elucidate the evolutionary history and divergence times of Neuropterida (Insecta: Holometabola). BMC Evolutionary Biology, 20, 64. https://doi.org/10.1186/s12862-020-01631-6
- Wang, H.C., Minh, B.Q., Susko, E. & Roger, A.J. (2018) Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Systematic Biology, 67, 216–235. https://doi.org/10.1093/sysbio/syx068
- Wang, Y.Y., Liu, X.Y., Garzón-Orduña, I.J., Winterton, S.L., Yan, Y., Aspöck, U., Aspöck, H. & Yang, D.Y. (2017) Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics, 33, 617–636. https://doi.org/10.1111/cla.12186
- Wang, Y.Y., Zhou, X.F., Wang, L.M., Liu, X.Y., Yang, D. & Rokas, A. (2019) Gene selection and evolutionary modelling affect phylogenomic inference of Neuropterida based on transcriptome data. International Journal of Molecular Sciences, 20, 1072. https://doi.org/10.3390/ijms20051072
- Winterton, S.L., Hardy, N.B. & Wiegmann, B.M. (2010) On wings of lace: phylogeny and Bayesian divergence time estimates of Neuropterida (Insecta) based on morphological and molecular data. Systematic Entomology, 35, 349–378. https://doi.org/10.1111/j.1365-3113.2010.00521.x
- Winterton, S.L., Lemmon, A., Gillung, J.P., Garzon, I.J., Badano, D., Bakkes, D.K., Breitkreuz, L.C.V., Engel, M.S., Lemmon, E.M., Liu, X.Y., Machado, R.J.P., Skevington, J.H. & Oswald, J.D. (2018) Evolution of lacewings and allied orders using anchored phylogenomics (Neuroptera, Megaloptera, Raphidioptera). Systematic Entomology, 43, 330–354. https://doi.org/10.1111/syen.12278
- Withycombe, C.L. (1925) XV. Some aspects of the biology and morphology of the Neuroptera. With special reference to the immature stages and their possible phylogenetic significance. Transactions of the Royal Entomological Society of London, 72, 303–411. https://doi.org/10.1111/j.1365-2311.1925.tb03362.x