Abstract
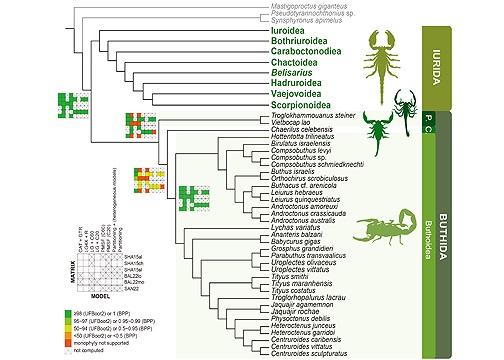
The evolutionary history of scorpions, one of the most charismatic groups of arthropods with over 2,500 described species, is a subject of great interest. Given their unique body plan and ancient fossil record, understanding their phylogeny is crucial. Recent genome-scale data have shown potential in elucidating interfamilial relationships within the scorpion tree of life; however, discrepancies between molecular datasets have also been observed. To address this, we explored the phylogenetic relationships of scorpions by synthesizing three previous phylogenomic studies using model comparison methods that account for among-site compositional heterogeneity. Our analyses indicate that under the infinite site-heterogeneous CAT and finite mixture models, the relationships within Scorpiones differ significantly from prevailing conclusions, particularly regarding the interfamilial relationships within Buthida. Our results show the following relationships: [Pseudochactoidea + [Buthoidea + Chaeriloidea]]. Our leave-one-out cross-validation analysis shows that the site-heterogeneous CAT-GTR+G model fits significantly better than the site-homogeneous model (LG) and finite mixture models (LG+C20, LG+C60), and it identifies contentious nodes in scorpion phylogeny. These analyses resolve a longstanding controversy in deep scorpion phylogeny and emphasize the significance of adequate modeling of compositional heterogeneity in reconstructing the deep phylogeny of scorpions.
References
- Ballesteros, J.A., Santibáñez-López, C.E., Baker, C.M., Benavides, L.R., Cunha, T.J., Gainett, G., Ontano, A.Z., Setton, E.V.W., Arango, C.P., Gavish-Regev, E., Harvey, M.S., Wheeler, W.C., Hormiga, G., Giribet, G. & Sharma, P.P. (2022) Comprehensive species sampling and sophisticated algorithmic approaches refute the monophyly of Arachnida. Molecular Biology and Evolution, 39, msac021. https://doi.org/10.1093/molbev/msac021
- Borowiec, M.L., Rabeling, C., Brady, S.G., Fisher, B.L., Schultz, T.R. & Ward, P.S. (2019) Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. Molecular Phylogenetics and Evolution, 134, 111–121. https://doi.org/10.1016/j.ympev.2019.01.024
- Bujaki, T. & Rodrigue, N. (2022) Bayesian cross-validation comparison of amino acid replacement models: contrasting profile mixtures, pairwise exchangeabilities, and gamma-distributed rates-across-sites. Journal of Molecular Evolution, 90, 468–475. https://doi.org/10.1007/s00239-022-10076-y
- Cai, C.Y., Tihelka, E., Liu, X.Y. & Engel, M.A. (2023) Improved modelling of compositional heterogeneity reconciles phylogenomic conflicts among lacewings. Palaeoentomology, 6 (1), 49–57. https://doi.org/10.11646/palaeoentomology.6.1.8
- Cai, C.Y. (2024) Ant backbone phylogeny resolved by modelling compositional heterogeneity among sites in genomic data. Communications Biology, 7, 106. https://doi.org/10.1038/s42003-024-05793-7
- Coddington, J.A., Giribet, G., Harvey, M.S., Prendini, L. & Walter, D.E. (2004) Arachnida. In: Cracraft, J. & Donoghue, M.J. (Eds), Assembling the tree of life. Oxford University Press, New York, 296–318. https://doi.org/10.1093/oso/9780195172348.003.0019
- Criscuolo, A. & Gribaldo, S. (2010) BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology, 10, 210. https://doi.org/10.1186/1471-2148-10-210
- Crotty, S.M., Minh, B.Q., Bean, N.G., Holland, B.R., Tuke, J., Jermiin, L.S. & von Haeseler, A. (2020) GHOST: Recovering historical signal from heterotachously evolved sequence alignments. Systematic Biology, 69, 249–264. https://doi.org/10.1093/sysbio/syz051
- de Carvalho, M.G., Maisey, J.G., Mendes, I.D. & de Souza Carvalho, I. (2023) Micro-tomographic analysis of a scorpion fossil from the Aptian Crato Formation of Northeastern Brazil. Cretaceous Research, 147, 105454. https://doi.org/10.1016/j.cretres.2022.105454
- Dunlop, J.A. & Selden, P.A. (2013) Scorpion fragments from the Silurian of Powys, Wales. Arachnology, 16, 27–32. https://doi.org/10.13156/arac.2013.16.1.27
- Dunlop, J.A. (2010) Geological history and phylogeny of Chelicerata. Arthropod Structure & Development, 39, 124–142. https://doi.org/10.1016/j.asd.2010.01.003
- Graham, M.R. & Fet, V. (2006) Serrula in retrospect: a historical look at scorpion literature (Scorpiones: Orthosterni). Euscorpius, 48, 1–9. https://doi.org/10.18590/euscorpius.2006.vol2006.iss48.1
- Harvey, M.S. (1992) The phylogeny and classification of the Pseudoscorpionida (Chelicerata: Arachnida). Invertebrate Systematics, 6, 1373–1435. https://doi.org/10.1071/IT9921373
- Hoff, C.C. (1949) The pseudoscorpions of Illinois. Illinois Natural History Survey Bulletin, 24, 407–498. https://doi.org/10.21900/j.inhs.v24.198
- Hoff, M., Orf, S., Riehm, B., Darriba, D. & Stamatakis, A. (2016) Does the choice of nucleotide substitution models matter topologically? BMC Bioinformatics, 17, 143. https://doi.org/10.1186/s12859-016-0985-x
- Howard, R.J., Edgecombe, G.D., Legg, D.A., Pisani, D. & Lozano-Fernandez, J. (2019) Exploring the evolution and terrestrialization of scorpions (Arachnida: Scorpiones) with rocks and clocks. Organisms Diversity & Evolution, 19, 71–86. https://doi.org/10.1007/s13127-019-00390-7
- Jeram, A.J. (1998) Phylogeny, classifications and evolution of Silurian and Devonian scorpions. In: Selden, P.A. (Ed.), Proceedings of the 17th European Colloquium of Arachnology, Edinburgh, UK. British Arachnological Society, Burnham Beeches, UK, 17–31.
- Kainer, D. & Lanfear, R. (2015) The effects of partitioning on phylogenetic inference. Molecular Biology and Evolution, 32, 1611–1627. https://doi.org/10.1093/molbev/msv026
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
- Kapli, P., Yang, Z.H. & Telford, M.J. (2020) Phylogenetic tree building in the genomic age. Nature Reviews Genetics, 21, 428–444. https://doi.org/10.1038/s41576-020-0233-0
- Kjellesvig-Waering, E.N. (1986) A restudy of the fossil Scorpionida of the world. Paleontological Research Institution, 55, 1–287.
- Kocot, K.M., Struck, T.H., Merkel, J., Waits, D.S., Todt, C., Brannock, P.M., Weese, D.A., Cannon, J.T., Moroz, L.L., Lieb, B. & Halanych, K.M. (2017) Phylogenomics of Lophotrochozoa with consideration of systematic error. Systematic Biology, 66, 256–282. https://doi.org/10.1093/sysbio/syw079
- Lamoral, B.H. (1980) A reappraisal of the suprageneric classification of recent scorpions and their zoogeography. In: Gruber, J. (Ed.), Verhandlungen. 8. Internationaler Arachnologen-Kongress abgehalten ander Universität für Bodenkultur Wien. 7–12 Juli, 1980. H. Egermann, Vienna, 439–444.
- Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. (2013) Phylobayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Systematic Biology, 62, 611–615. https://doi.org/10.1093/sysbio/syt022
- Lartillot, N. (2023) Identifying the best approximating model in Bayesian phylogenetics: Bayes factors, cross-validation or wAIC? Systematic Biology, 72, 616–638. https://doi.org/10.1093/sysbio/syad004
- Laumer, C.E., Fernández, R., Lemer, S., Combosch, D., Kocot, K.M., Riesgo, A., Andrade, S.C.S., Sterrer, W., Sørensen, M.V. & Giribet, G. (2019) Revisiting metazoan phylogeny with genomic sampling of all phyla. Proceedings of the Royal Society B: Biological Sciences, 286, 20190831. https://doi.org/10.1098/rspb.2019.0831
- Le, S.Q., Dang, C.C. & Gascuel, O. (2012) Modeling protein evolution with several amino acid replacement matrices depending on site rates. Molecular Biology and Evolution, 29, 2921–2936. https://doi.org/10.1093/molbev/mss112
- Legg, D.A., Garwood, R.J., Dunlop, J.A. & Sutton, M.D. (2012) A taxonomic revision of orthosternous scorpions from the English Coal-Measures aided by X-ray micro-tomography (XMT). Palaeontologia Electronica, 15, 1–16. https://doi.org/10.26879/253
- Lourenço, W.R. (2001) A remarkable scorpion fossil from the amber of Lebanon. Implications for the phylogeny of Buthoidea. Comptes Rendus de l’Académie des Sciences—Series IIA—Earth and Planetary Science, 332, 641–646. https://doi.org/10.1016/S1251-8050(01)01583-X
- Lozano-Fernandez, J. (2022) A practical guide to design and assess a phylogenomic study. Genome Biology and Evolution, 14, evac129. https://doi.org/10.1093/gbe/evac129
- Maddison, W.P. & Hedin, M.C. (2003) Jumping spider phylogeny (Araneae: Salticidae). Invertebrate Systematics, 17, 529–549. https://doi.org/10.3897/zookeys.440.7891
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Misof, B., Liu, S., Meusemann, K., Peters, R.S., Donath, A., Mayer, C., Frandsen, P.B., Ware, J., Flouri, T., Beutel, R.G., Niehuis, O., Petersen, M., Izquierdo-Carrasco, F., Wappler, T., Rust, J., Aberer, A.J., Aspöck, U., Aspöck, H., Bartel, D., Blanke, A., Berger, S., Böhm, A., Buckley, T.R., Calcott, B., Chen, J., Friedrich, F., Fukui, M., Fujita, M., Greve, C., Grobe, P., Gu, S., Huang, Y., Jermiin, L.S., Kawahara, A.Y., Krogmann, L., Kubiak, M., Lanfear, R., Letsch, H., Li, Y., Li, Z., Li, J., Lu, H., Machida, R., Mashimo, Y., Kapli, P., McKenna, D.D., Meng, G., Nakagaki, Y., Navarrete-Heredia, J.L., Ott, M., Ou, Y., Pass, G., Podsiadlowski, L., Pohl, H., Reumont, B.M. von, Schütte, K., Sekiya, K., Shimizu, S., Slipinski, A., Stamatakis, A., Song, W., Su, X., Szucsich, N.U., Tan, M., Tan, X., Tang, M., Tang, J., Timelthaler, G., Tomizuka, S., Trautwein, M., Tong, X., uchifune, T., Walzl, M.G., Wiegmann, B.M., Wilbrandt, J., Wipfler, B., Wong, T.K.F., Wu, Q., Wu, G., Xie, Y., Yang, S., Yang, Q., yeates, D.K., Yoshizawa, K., Zhang, Q., Zhang, R., Zhang, W., Zhang, Y., Zhao, J., Zhou, C., Zhou, L., Ziesmann, T., Zou, S., Li, Y., Xu, X., Zhang, Y., Yang, H., Wang, J., Wang, J., Kjer, K.M. & Zhou, X. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science, 346, 763–767. https://doi.org/10.1126/science.1257570
- Polis, G.A. (1990) The biology of scorpions. Stanford University Press, Stanford, California, 587 pp.
- Prendini, L., Volschenk, E., Maaliki, S. & Gromov, A.V. (2006) A ‘living fossil’ from Central Asia: the morphology of Pseudochactas ovchinnikovi Gromov, 1998 (Scorpiones: Pseudochactidae), with comments on its phylogenetic position. Zoologischer Anzeiger—A Journal of Comparative Zoology, 245, 211–248. https://doi.org/10.1016/j.jcz.2006.07.001
- Rowland, J.M. (1975) Classification, phylogeny and zoogeography of the American arachnids of the order Schizomida. Doctoral thesis, Texas Tech University, 415 pp.
- Santibáñez-López, C.E., Aharon, S., Ballesteros, J.A., Gainett, G., Baker, C.M., González-Santillán, E., Harvey, M.S., Hassan, M.K., Almaaty, A.H.A, Aldeyarbi, S.M., Monod, L., Ojanguren-Affilastro, A., Pinto-da-Rocha, R., Zvik, Y., Gavish-Regev, E. & Sharma, P.P. (2022) Phylogenomics of scorpions reveal contemporaneous diversification of scorpion mammalian predators and mammal-active sodium channel toxins. Systematic Biology, 71, 1281–1289. https://doi.org/10.1093/sysbio/syac021
- Sharma, P.P., Baker, C.M., Cosgrove, J.G., Johnson, J.E., Oberski, J.T., Raven, R.J., Harvey, M.S., Boyer, S.L. & Giribet, G. (2018) A revised dated phylogeny of scorpions: Phylogenomic support for ancient divergence of the temperate Gondwanan family Bothriuridae. Molecular Phylogenetics and Evolution, 122, 37–45. https://doi.org/10.1016/j.ympev.2018.01.003
- Sharma, P.P., Fernández, R., Esposito, L.A., González-Santillán, E. & Monod, L. (2015) Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal. Proceedings of The Royal Society B-biological Sciences, 282, 20142953. https://doi.org/10.1098/rspb.2014.2953
- Simion, P., Philippe, H., Baurain, D., Jager, M., Richter, D.J., Franco, A.D., Roure, B., Satoh, N., Quéinnec, E., Ereskovsky, A., Lapébie, P., Corre, E., Delsuc, F., King, N., Wörheide, G. & Manuel, M. (2017) A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Current Biology, 27, 958–967. https://doi.org/10.1016/j.cub.2017.02.031
- Sissom, W.D. (1990) Systematics, biogeography, and paleontology. In: Polis, G.A. (Ed.), The biology of scorpions. Stanford University Press, Stanford, California, 64–160.
- Siu-Ting, K., Torres-Sanchez, M., Mauro, D.S., Wilcockson, D., Wilkinson, M., Pisani, D., O’Connell, M.J. & Creevey, C.J. (2019) Inadvertent paralog inclusion drives artifactual topologies and timetree estimates in phylogenomics. Molecular Biology and Evolution, 36, 1344–1356. https://doi.org/10.1093/molbev/msz067
- Smith, B.T., Mauck, W.M., Benz, B.W. & Andersen, M.J. (2020) Uneven missing data skew phylogenomic relationships within the lories and lorikeets. Genome Biology and Evolution, 12, 1131–1147. https://doi.org/10.1093/gbe/evaa113
- Soleglad, M.E. & Fet, V. (2003a) High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius, 11, 1–57. https://doi.org/10.18590/euscorpius.2003.vol2003.iss11.1
- Soleglad, M.E. & Fet, V. (2003b) The scorpion sternum: structure and phylogeny (Scorpiones: Orthosterni). Euscorpius, 5, 1–34. https://doi.org/10.18590/euscorpius.2003.vol2003.iss5.1
- Stockwell, S.A. (1989) Revision of the phylogeny and higher classification of scorpions (Chelicerata). PhD thesis, University of Berkeley, Berkeley, California. University Microfilms International, Ann Arbor, Michigan, 319 pp.
- Strimmer, K. & von Haeseler, A. (1997) Likelihood-mapping: A simple method to visualize phylogenetic. Proceedings of National Academy of Sciences, 94, 6815–6819. https://doi.org/10.1073/pnas.94.13.6815
- Tihelka, E., Cai, C.Y., Giacomelli, M., Lozano-Fernandez, J., Rota-Stabelli, O., Huang, D.Y., Engel, M.S., Donoghue, P.C. & Pisani, D. (2021) The evolution of insect biodiversity. Current Biology, 31, R1299–R1311. https://doi.org/10.1016/j.cub.2021.08.057
- Waddington, J., Rudkin, D.M. & Dunlop, J.A. (2015) A new mid-Silurian aquatic scorpion-one step closer to land? Biology Letters, 11, 20140815. https://doi.org/10.1098/rsbl.2014.0815
- Wang, H.C., Minh, B.Q., Susko, E. & Roger, A.J. (2018) Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Systematic Biology, 67, 216–235. https://doi.org/10.1093/sysbio/syx068
- Wang, Y., Li, Y.D., Wang, S., Tihelka, E., Engel, M.S. & Cai, C.Y. (2024) Modeling compositional heterogeneity resolves deep phylogeny of flowering plants. Plant Diversity. https://doi.org/10.1016/j.pld.2024.07.007
- Young, A.D. & Gillung, J.P. (2020) Phylogenomics—principles, opportunities and pitfalls of big-data phylogenetics. Systematic Entomology, 45, 225–247. https://doi.org/10.1111/syen.12406