Abstract
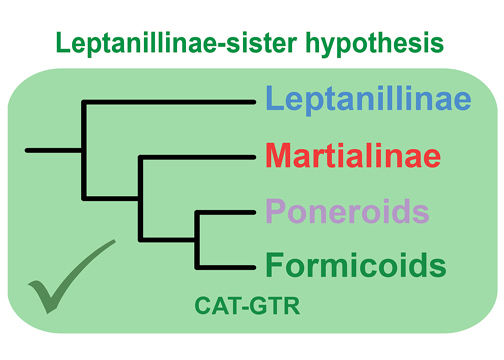
Understanding the early evolution of ants has been hindered by conflicting phylogenetic hypotheses and methodological inconsistencies across studies. In Cai (2024), I reanalyzed both Sanger-sequencing and genome-scale datasets of ants using rigorous model comparison and methods that account for among-site compositional heterogeneity to identify the sources of phylogenetic conflict. The results showed that the 11-loci datasets in Borowiec et al. (2019) failed to resolve deep ant relationships and could not determine the position of Martialis heureka. Analyses of the genome-scale data further revealed that the placement of key lineages depends strongly on model fit. Bayesian cross-validation and posterior predictive assessments demonstrated that the infinite mixture CAT-GTR+G4 model substantially outperforms empirical finite mixture models, providing robust support for the Leptanillinae-sister hypothesis. Criticisms by Boudinot & Lieberman (2025) regarding the study design, model choice, and convergence assessments stem from misinterpretations of the analytical framework. The matrices in Cai (2024) were explicitly designed to test model performance under controlled subsampling and filtering schemes, and all analyses showed consistent results across datasets. The findings reaffirm that accurately modelling among-site compositional heterogeneity is essential for resolving the backbone phylogeny of ants, and that under the best-fitting models, Martialis heureka occupies a well-supported position as sister to all non-leptanilline ants.
References
- Borowiec, M.L., Rabeling, C., Brady, S.G., Fisher, B.L., Schultz, T.R. & Ward, P.S. (2019) Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. Molecular Phylogenetics and Evolution, 134, 111–121. https://doi.org/10.1016/j.ympev.2019.01.024
- Boudinot, B.E. & Lieberman, Z.E. (2025) Ant phylogeny is not resolved by the application of site heterogeneous models. Communications Biology, 8, 1458. https://doi.org/10.1038/s42003-025-07849-8
- Boudinot, B.E., Khouri, Z., Richter, A., Griebenow, Z.H., van de Kamp, T., Perrichot, V. & Barden, P. (2022) Evolution and systematics of the Aculeata and kin (Hymenoptera), with emphasis on the ants (Formicoidea: †@@@idae fam. nov., Formicidae). bioRxiv, preprint. https://doi.org/10.1101/2022.02.20.480183
- Boudinot, B.E., Fikáček, M., Lieberman, Z.E., Kusy, D., Bocak, L., McKenna, D.D. & Beutel, R.G. (2022) Systematic bias and the phylogeny of Coleoptera—A response to Cai et al. (2022) following the responses to Cai et al. (2020). Systematic Entomology, 48, 223–232. https://doi.org/10.1111/syen.12570
- Bujaki, T. & Rodrigue, N. (2022) Bayesian cross-validation comparison of amino acid replacement models: contrasting profile mixtures, pairwise exchangeabilities, and gamma-distributed rates-across-sites. Journal of Molecular Evolution, 90, 468–475. https://doi.org/10.1007/s00239-022-10076-y
- Cai, C.Y. (2024) Ant backbone phylogeny resolved by modelling compositional heterogeneity among sites in genomic data. Communications Biology, 7, 106. https://doi.org/10.1038/s42003-024-05793-7
- Cai, C.Y., Tihelka, E., Pisani, D. & Donoghue, P.C. (2024) Resolving incongruences in insect phylogenomics: a reply to Boudinot et al. (2023). Palaeoentomology, 7 (2), 176–183. https://doi.org/10.11646/palaeoentomology.7.2.2
- Criscuolo, A. & Gribaldo, S. (2010) BMGE (Block Mapping and Gathering with Entropy): selection of phylogenetic informative regions from multiple sequence alignments. BMC Evolutionary Biology, 10, 210. https://doi.org/10.1186/1471-2148-10-210
- Crotty, S.M., Minh, B.Q., Bean, N.G., Holland, B.R., Tuke, J., Jermiin, L.S. & Haeseler, A.V. (2020) GHOST: recovering historical signal from heterotachously evolved sequence alignments. Systematic Biology, 69, 249–264. https://doi.org/10.1093/sysbio/syz051
- Cunha, T.J., Reimer, J.D. & Giribet, G. (2022) Investigating sources of conflict in deep phylogenomics of vetigastropod snails. Systematic Biology, 71, 1009–1022. https://doi.org/10.1093/sysbio/syab071
- Fabreti, L.G. & Höhna, S. (2022) Bayesian inference of phylogeny is robust to substitution model over-parameterization. bioRxiv, preprint. https://doi.org/10.1101/2022.02.17.480861
- Feuda, R., Dohrmann, M., Pett, W., Philippe, H., Rota-Stabelli, O., Lartillot, N., Wörheide, G. & Pisani, D. (2017) Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Current Biology, 27, 3864–3870. https://doi.org/10.1016/j.cub.2017.11.008
- Fleming, J.F., Valero‐Gracia, A. & Struck, T.H. (2023) Identifying and addressing methodological incongruence in phylogenomics: A review. Evolutionary Applications, 16, 1087–1104. https://doi.org/10.1111/eva.13565
- Giacomelli, M., Rossi, M.E., Lozano-Fernandez, J., Feuda, R. & Pisani, D. (2022) Resolving tricky nodes in the tree of life through amino acid recoding. iScience, 25, 105594. https://doi.org/10.1016/j.isci.2022.105594
- Janouškovec, J., Tikhonenkov, D.V., Burki, F., Howe, A.T., Rohwer, F.L., Mylnikov, A.P. & Keeling, P.J. (2017) A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Current Biology, 27, 3717–3724. https://doi.org/10.1016/j.cub.2017.10.051
- Kapli, P., Flouri, T. & Telford, M.J. (2021) Systematic errors in phylogenetic trees. Current Biology, 31, R59–R64. https://doi.org/10.1016/j.cub.2020.11.043
- Kapli, P., Yang, Z. & Telford, M.J. (2020) Phylogenetic tree building in the genomic age. Nature Reviews Genetics, 21 (7), 428–444. https://doi.org/10.1038/s41576-020-0233-0
- Lartillot, N. (2020) PhyloBayes: Bayesian phylogenetics using site-heterogeneous models. In: Scornavacca, C., Delsuc, F. & Galtier, N. (Eds), Phylogenetics in the genomic era, pp. 1.5:1–1.5:16. No commercial publisher, author’s open access book. https://hal.science/PGE
- Lartillot, N. (2023) Identifying the best approximating model in Bayesian phylogenetics: Bayes factors, cross-validation or wAIC? Systematic Biology, 72, 616–638. https://doi.org/10.1093/sysbio/syad004
- Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. (2013) PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Systematic Biology, 62, 611–615. https://doi.org/10.1093/sysbio/syt022
- Laumer, C.E., Gruber-Vodicka, H.R., Hadfield, M.G., Pearse, V.B., Riesgo, A., Marioni, J.C. & Giribet, G. (2018) Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. eLife, 7, e36278. https://doi.org/10.7554/eLife.36278
- Le, S.Q., Dang, C.C. & Gascuel, O. (2012) Modeling protein evolution with several amino acid replacement matrices depending on site rates. Molecular Biology and Evolution, 29, 2921–2936. https://doi.org/10.1093/molbev/mss112
- Li, Y.D., Engel, M.S., Tihelka, E. & Cai, C.Y. (2023) Phylogenomics of weevils revisited: data curation and modelling compositional heterogeneity. Biology Letters, 19, 20230307. https://doi.org/10.1098/rsbl.2023.0307
- Liu, Y., Cox, C.J., Wang, W. & Goffinet, B. (2014) Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Systematic Biology, 63 (6), 862–878. https://doi.org/10.1093/sysbio/syu049
- Lozano-Fernandez, J., Tanner, A.R., Giacomelli, M., Carton, R., Vinther, J., Edgecombe, G.D. & Pisani, D. (2019a) Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nature Communications, 10, 2295. https://doi.org/10.1038/s41467-019-10244-7
- Lozano-Fernandez, J., Giacomelli, M., Fleming, J.F., Chen, A., Vinther, J., Thomsen, P.F., Glenner, H., Palero, F., Legg, D.A., Iliffe, T.M. & Pisani, D. (2019b) Pancrustacean evolution illuminated by taxon-rich genomic-scale data sets with an expanded remipede sampling. Genome Biology and Evolution, 11, 2055–2070. https://doi.org/10.1093/gbe/evz097
- Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T.J. (2018) Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature, 557, 101–105. https://doi.org/10.1038/s41586-018-0059-5
- Mulhair, P.O., McCarthy, C.G., Siu-Ting, K., Creevey, C.J. & O’Connell, M.J. (2022) Filtering artifactual signal increases support for Xenacoelomorpha and Ambulacraria sister relationship in the animal tree of life. Current Biology, 32, 5180–5188. https://doi.org/10.1016/j.cub.2022.10.036
- Ochoa de Alda, J.A., Esteban, R., Diago, M.L. & Houmard, J. (2014) The plastid ancestor originated among one of the major cyanobacterial lineages. Nature Communications, 5, 4937. https://doi.org/10.1038/ncomms5937
- Philippe, H., Poustka, A.J., Chiodin, M., Hoff, K.J., Dessimoz, C., Tomiczek, B., Schiffer, P.H., Müller, S., Domman, D., Horn, M., Kuhl, H., Timmermann, B., Satoh, N., Hikosaka-Katayama, T., Nakano, H., Rowe, M.L., Elphick, M.R., Thomas-Chollier, M., Hankeln, T., Mertes, F., Wallberg, A., Rast, J.P., Copley, R.R., Martinez, P. & Telford, M.J. (2019) Mitigating anticipated effects of systematic errors supports sister-group relationship between Xenacoelomorpha and Ambulacraria. Current Biology, 29, 1818–1826. https://doi.org/10.1016/j.cub.2019.04.009
- Romiguier, J., Borowiec, M.L., Weyna, A., Helleu, Q., Loire, E., La Mendola, C., Rabeling, C., Fisher, B.L., Ward, P.S. & Keller, L. (2022) Ant phylogenomics reveals a natural selection hotspot preceding the origin of complex eusociality. Current Biology, 32, 2942–2947. https://doi.org/10.1016/j.cub.2022.05.001
- Schrempf, D., Lartillot, N. & Szöllősi, G. (2020) Scalable empirical mixture models that account for across-site compositional heterogeneity. Molecular Biology and Evolution, 37 (12), 3616–3631. https://doi.org/10.1093/molbev/msaa145
- Strassert, J.F. & Monaghan, M.T. (2022) Phylogenomic insights into the early diversification of fungi. Current Biology, 32, 3628–3635. https://doi.org/10.1016/j.cub.2021.08.057
- Tihelka, E., Cai, C.Y., Giacomelli, M., Lozano-Fernandez, J., Rota-Stabelli, O., Huang, D.Y., Engel, M.S., Donoghue, P.C. & Pisani, D. (2021) The evolution of insect biodiversity. Current Biology, 31, R1299–R1311. https://doi.org/10.1016/j.cub.2021.08.057
- Wang, H.C., Minh, B.Q., Susko, E. & Roger, A.J. (2018) Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Systematic Biology, 67, 216–235. https://doi.org/10.1093/sysbio/syx068
- Whelan, N.V. & Halanych, K.M. (2017) Who let the CAT out of the bag? Accurately dealing with substitutional heterogeneity in phylogenomic analyses. Systematic Biology, 66 (2), 232–255. https://doi.org/10.1093/sysbio/syw084
- Zaremba-Niedzwiedzka, K., Caceres, E.F., Saw, J.H., Bäckström, D., Juzokaite, L., Vancaester, E., Seitz, K.W., Anantharaman, K., Starnawski, P., Kjeldsen, K.U., Stott, M.B., Nunoura, T., Banfield, J.F., Schramm, A., Baker, B.J., Spang, A. & Ettema, T.J.G. (2017) Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature, 541, 353–358. https://doi.org/10.1038/nature21031