Abstract
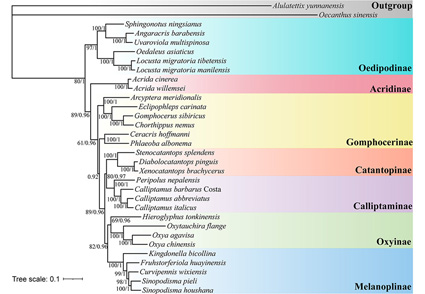
Calliptamus barbarus Costa is a species in genus Calliptamus, belonging to the Calliptaminae, Acrididae, Orthoptera. In the present study, the mitogenome of C. barbarous was determined and annotated to better identify C. barbarous and other related species. The mitogenome was 15,686 bp in length and encoded 37 genes, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and a control region. The nucleotide composition analysis of the mitogenome showed a strong bias of A/T, with A+T content of 72.7%, being similar to other reported mitogenomes. All the 13-PCGs used typical start codons of ATN and stop codons of TAA/G. All of the transfer RNA genes had typical clover-leaf structure except the missing dihydrouidine (DHU) arm of tRNASer1. Interestingly, we observed two overlapped but conserved regions between ATP8 and ATP6 genes, ND4 and ND4L genes. The results of Ka/Ks implied that CytB is more conservative than COX1. Phylogenetic analyses based on the 13-PCGs from 30 species indicated strong support for the monophyly of Acridoidea and represented the main topology as follows: (Oedipodinae+ (Acridinae+ (Gomphocerinae +((Calliptaminae+Catantopinae) +(Oxyinae+Melanoplinae))))). The phylogenetic relationship indicated that the molecular taxonomy of C. barbarous was consistent with the current morphological classification. The data in the present study will further enrich the mitogenome database of Acridoidea and provided useful clues for further research of C. barbarous on the mitogenome evolution and diversification of Calliptamus.
References
Bernt, M., Braband, A., Middendorf, M., Misof, B., Rota-Stabelli, O. & Stadler, P.F. (2013) Bioinformatics methods for the comparative analysis of metazoan mitochondrial genome sequences. Molecular Phylogenetics and Evolution, 69, 320−327. https://doi.org/10.1016/j.ympev.2012.09.019
Bernt, M., Donath, A., Juhling, F., Externbrink, F., Florentz, C., Fritzsch, G., Putz, J., Middendorf, M. & Stadler, P.F. (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69, 313−319. https://doi.org/10.1016/j.ympev.2012.08.023
Cameron, S.L. (2014) How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Systematic Entomology, 39, 400−411. https://doi.org/10.1111/syen.12071
Cameron, S.L. & Whiting, M.F. (2007) Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome, 50, 188−202. https://doi.org/10.1139/g06-148
Cao, S.S., Yu, W.W., Sun, M. & Du, Y.Z. (2014) Characterization of the complete mitochondrial genome of Tryporyza incertulas, in comparison with seven other Pyraloidea moths. Gene, 533, 356−365. https://doi.org/10.1016/j.gene.2013.07.072
Chang, H.H., Qiu, Z.Y., Yuan, H., Wang, X.Y., Li, X.J., Sun, H.M., Guo, X.Q., Lu, Y.C., Feng, X.L., Majid, M. & Huang, Y. (2020) Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Molecular Phylogenetics and Evolution, 145, 106734. https://doi.org/10.1016/j.ympev.2020.106734
da Silva, F.S., Cruz, A.C.R., de Almeida Medeiros, D.B., da Silva, S.P., Nunes, M.R.T., Martins, L.C., Chiang, J.O., da Silva, L.P., Cunha, G.M., de Araujo, R.F., de Oliveira Monteiro, H.A. & Nunes Neto, J.P. (2020) Mitochondrial genome sequencing and phylogeny of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis (Diptera: Culicidae). Scientific Reports, 10, 16948. https://doi.org/10.1038/s41598-020-73790-x
Deng, J.L., Yu, Z.Q, Ning, J., Wang, H., Lin, X.D. & Liu, X.L. (2020) Complete mitochondrial genome and phylogenetic analysis of a Chorthippus fallax (Zuboxsky) isolated in rangeland of northwest region of China. Mitochondrial DNA part B, 5, 61−62. https://doi.org/10.1080/23802359.2019.1695549
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059−3066. https://doi.org/10.1093/nar/gkf436
Kerpedjiev, P., Hammer, S. & Hofacker, I.L. (2015) Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics, 31, 3377−3379. https://doi.org/10.1093/bioinformatics/btv372
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evoluti nary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35, 1547−1549. https://doi.org/10.1093/molbev/msy096
Li, F., Lv, Y.Y., Wen, Z.Y., Bian, C., Zhang, X.H., Guo, S.T., Shi, Q. & Li, D.Q. (2021) The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders. BMC Ecology and Evolution, 21, 72. https://doi.org/10.1186/s12862-021-01803-y
Li, X.J., Zhi, Y.C., Liu, G.J., Yin, X.C. & Zhang, D.C. (2015) The complete mitochondrial genome of Asiotmethis jubatus (Uvarov, 1926) (Orthoptera: Acridoidea: Pamphagidae). Mitochondrial DNA, 26 (5), 785−786. https://doi.org/10.3109/19401736.2013.855752
Lowe, T.M. & Eddy, S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25, 955−964. https://doi.org/10.1093/nar/25.5.955
Malyarchuk, B., Derenko, M., Berman, D., Perkova, M., Grzybowski, T., Lejrikh, A. & Bulakhova, N. (2010) Phylogeography and molecular adaptation of Siberian salamander Salamandrella keyserlingii based on mitochondrial DNA variation. Molecular Phylogenetics and Evolution, 56, 562−571. https://doi.org/10.1016/j.ympev.2010.04.005
Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268−274. https://doi.org/10.1093/molbev/msu300
Perna, N.T. & Kocher, T.D. (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution, 41, 353−358. https://doi.org/10.1007/BF01215182
Qian, Y.H., Wu, H.Y., Ji, X.Y., Yu, W.W. & Du, Y.Z. (2014) Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of plecoptera based on mitogenomes. PLoS ONE, 9, e86328. https://doi.org/10.1371/journal.pone.0086328
Ranwez, V., Harispe, S., Delsuc, F. & Douzery, E.J. (2011) MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE, 6, e22594. https://doi.org/10.1371/journal.pone.0022594
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539−542. https://doi.org/10.1093/sysbio/sys029
Rouibah, M., Lopez-Lope, A., Presa, J.J. & Doumandji, S. (2016) A molecular phylogenetic and phylogeographic study of two forms of Calliptamus barbarus (Costa 1836) (Orthoptera: Acrididae, Calliptaminae) from two regions of Algeria. Annales de La Societe Entomologique de Frace, 52, 77−87. https://doi.org/10.1080/00379271.2016.1188329
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J.C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S.E. & Sanchez-Gracia, A. (2017) DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Molecular Biology and Evolution, 34, 3299−3302. https://doi.org/10.1093/molbev/msx248
Salvato, P., Simonato, M., Battisti, A. & Negrisolo, E. (2008) The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics, 9, 331. https://doi.org/10.1186/1471-2164-9-331
Sergeev, M.G. (1992) Distribution Patterns of Orthoptera in North and Central Asia. Journal of Orthoptera Research, 76, 72−78. https://doi.org/10.2307/3503557
Simon, C. (1991) Molecular systematics at the species boundary: Exploiting conserved and variable regions of the mitochondrial genome of animals via direct sequencing from amplified DNA. In: Hewitt, G.M, Johnston, A.W.B. & Young, J.P.W. (Eds.), Molecular techniques in taxonomy. NATO ASI series, Springer-Verlag, Berlin, pp. 33−71. https://doi.org/10.1007/978-3-642-83962-7_4
Song, N., Li, X.X., Zhai, Q., Bozdogan, H. & Yin, X.M. (2019) The Mitochondrial Genomes of Neuropteridan Insects and Implications for the Phylogeny of Neuroptera. Genes, 10, 2. https://doi.org/10.3390/genes10020108
Wang, Q.Q. & Tang, G.H. (2017) Genomic and phylogenetic analysis of the complete mitochondrial DNA sequence of walnut leaf pest Paleosepharia posticata (Coleoptera: Chrysomeloidea). Journal of Asia-Pacific Entomology, 20, 840−853. https://doi.org/10.1016/j.aspen.2017.05.010
Xia, J.H., Li, H.L., Zhang, Y., Meng, Z.N. & Lin, H.R. (2018) Identifying selectively important amino acid positions associated with alternative habitat environments in fish mitochondrial genomes. Mitochondrial DNA Part A, 29, 511−524. https://doi.org/10.1080/24701394.2017.1315567
Xiao, B., Chen, W., Hu, C.C. & Jiang, G.F. (2012) Complete mitochondrial genome of the groundhopper Alulatettix yunnanensis (Insecta: Orthoptera: Tetrigoidea). Mitochondrial DNA, 23, 286−287. https://doi.org/10.3109/19401736.2012.674122
Ye, F., Liu, T., King, S.D. & You, P. (2015) Mitochondrial genomes of two phlebotomine sand flies, Phlebotomus chinensis and Phlebotomus papatasi (Diptera: Nematocera), the first representatives from the family Psychodidae. Parasites & Vectors, 8, 472. https://doi.org/10.1186/s13071-015-1081-1
Yin, X.C., Li, X.J., Wang, W.Q., Yin, H., Cao, C.Q., Ye, B.H. & Yin, Z. (2008) Phylogenetic analyses of some genera in Oedipodidae (Orthoptera: Acridoidea) based on 16S mitochondrial partial gene sequences. Insect Science, 15 (5), 471−476. https://doi.org/10.1111/j.1744-7917.2008.00235.x
Yu, Y.Q., Ma, W.M., Yang, W.J. & Yang, J.S. (2014) The complete mitogenome of the lined shore crab Pachygrapsus crassipes Randall 1840 (Crustacea: Decapoda: Grapsidae). Mitochondrial DNA, 25, 263−264. https://doi.org/10.3109/19401736.2013.800497
Zhang, C.Y. & Huang, Y. (2008) Complete mitochondrial genome of Oxya chinensis (Orthoptera, Acridoidea). Acta Biochimica et Biophysica Sinica, 40, 7−18. https://doi.org/10.1111/j.1745-7270.2008.00375.x
Zhang, D., Gao, F., Jakovlic, I., Zou, H, Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348−355. https://doi.org/10.1111/1755-0998.13096
Zhang, L., Lecoq, M., Latchininsky, A. & Hunter, D. (2019) Locust and Grasshopper Management. Annual Review of Entomology, 64, 15−34. https://doi.org/10.1146/annurev-ento-011118-112500