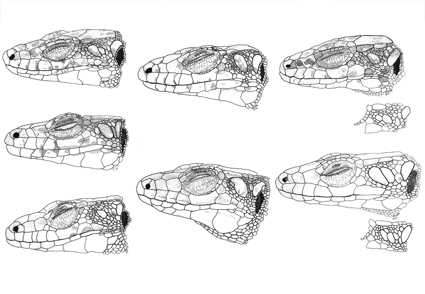
Abstract
Revealing biodiversity allows the accurate determination of the underlying causes of many biological processes such as speciation and hybridization. These processes contain many complex patterns, especially in areas with high species diversity. As two of the prominent zoogeographic areas, Anatolia and Caucasus are also home to the genus Darevskia, which has a complex morphological structure and parthenogenetic speciation. Darevskia valentini and D. rudis are two largely distributed taxa of this genus, both of which have a controversial taxonomic delimitation. Here we performed both a highly detailed morphological comparison and a molecular evaluation for the populations in both species groups. The most comprehensive taxonomic revision of this complex was carried out to determine the cases where the data obtained were compatible or not with each approach. As a result of the obtained outputs, it seems that D. spitzenbergerae stat. nov., D. mirabilis stat. nov. and D. obscura stat. nov. should be accepted as the species level, this later with subspecies D. o. bischoffi comb. nov. and D. o. macromaculata comb. nov.. Also, we propose two new taxa: D. josefschmidtleri sp. nov. and D. spitzenbergerae wernermayeri ssp. nov.. It has also been shown that “lantzicyreni” subspecies belong to D. rudis instead of D. valentini. The extensive revision has contributed to subsequent studies to more accurately understand the past histories of species in the genus Darevskia.
References
Ahmadzadeh, F., Flecks, M., Carretero, M.A., Mozaffari, O., Böhme, W., Harris, D.J., Freitas, S. & Rödder, D. (2013) Cryptic speciation patterns in Iranian Rock Lizards uncovered by integrative taxonomy. Plos One, 8 (12), 1–17. https://doi.org/10.1371/journal.pone.0080563
Arnold, E.N. (1973) Relationships of the Palaearctic lizards assigned to the genera Lacerta, Algyroides and Psammodromus (Reptilia: Lacertidae). Bulletin of the British Museum (Natural History) Zoology, London, 25 (8), 289–366.
Arnold, E.N., Arribas, O.J. & Carranza, S. (2007) Systematics of the Palaearctic and Oriental lizard tribe Lacertini (Squamata: Lacertidae: Lacertinae), with descriptions of eight new genera. Zootaxa, 1430 (1), 1–86. https://doi.org/10.11646/zootaxa.1430.1.1
Arribas, O.J. (1998) Osteology of the Pyrenaean Mountain Lizards and comparison with other species of the collective genus Archaeolacerta Mertens, 1921 s.l. from Europe and Asia Minor (Squamata: Lacertidae). Herpetozoa, Wien, 11 (1/2), 47–70.
Arribas, O.J. (1999) Phylogeny and relationships of the mountain lizards of Europe and Near East (Archaeolacerta Mertens, 1921, Sensu lato) and their relationships among the Eurasian lacertid radiation. Russian Journal of Herpetology, 6 (1), 1–22.
Arribas, O., Carranza, S. & Odierna, G. (2006) Description of a new endemic species of mountain lizard from Northwestern Spain: Iberolacerta galani sp. nov. (Squamata: Lacertidae). Zootaxa, 1240 (1), 1–55. https://doi.org/10.11646/zootaxa.1240.1.1
Arribas, O.J. (2010) Intraspecific variability of the Carpetane Lizard (Iberolacerta cyreni [Müller & Hellmich, 1937]) (Squamata: Lacertidae), with special reference to the unstudied peripheral populations from the Sierras de Avila (Paramera, Serrota and Villafranca). Bonn Zoological Bulletin, 57 (2), 197–210.
Arribas, O., Ilgaz, Ç., Kumlutaş, Y., Durmuş, S.H., Avcı, A. & Üzüm, N. (2013) External morphology and osteology of Darevskia rudis (Bedriaga, 1886), with a taxonomic revision of the Pontic and Small-Caucasus populations (Squamata: Lacertidae). Zootaxa, 3626 (4), 401–428. https://doi.org/10.11646/zootaxa.3626.4.1
Arribas, O., Candan, K., Kurnaz, M., Kumlutaş, Y., Yıldırım-Caynak, E. & Ilgaz, Ç. (2022) A new cryptic species of the Darevskia parvula group from NE Anatolia (Squamata, Lacertidae). Organisms Diversity & Evolution, 22, 475–490. https://doi.org/10.1007/s13127-022-00540-4
Baran, İ., Avcı, A., Kumlutaş, Y., Olgun, K. & Ilgaz Ç. (2021) Türkiye Amfibi ve Sürüngenleri, Palme Yayınevi, 2028, 223 pp.
Başoğlu, M. & Baran, I. (1977) Türkiye Sürüngenleri, Kısım I, Kaplumbağa ve Kertenkeleler [Turkish Reptiles, Part I, Turtles and Lizards]. Ege Üniversitesi Kitaplar Serisi, 76, 1–219.
Blackith, R.E. & Reyment, R.A. (1971) Multivariate morphometrics. Academic Press, London and New York, 412 pp.
Bouckaert, R., Vaughan, T.G., Barido-Sottani, J., Duchene, S., Fourment, M., Gavryushkina, A., Heled, J., Jones, G., Kuhnert, D., De Maio, N., Matschiner, M., Mendes, F.K., Muller, N. F., Ogilvie, H.A., Du Plessis, L., Popinga, A., Rambaut, A., Rasmussen, D., Siveroni, I., Suchard, M.A., Wu, C.H., Xie, D., Zhang, C., Stadler, T. & Drummond, A.J. (2019) BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 15 (4), e1006650. https://doi.org/10.1371/journal.pcbi.1006650
Böhme, W. & Budak, A. (1977) Uber die rudis-Gruppe des Lacerta saxicola-Komplexes in der Turkei, II (Reptilia: Sauria: Lacertidae). Salamandra, 13 (3/4), 141–149.
Busschau, T., Conradie, W. & Daniels, S.R. (2019) Evidence for cryptic diversification in a rupicolous forestdwelling gecko (Gekkonidae: Afroedura pondolia) from a biodiversity hotspot. Molecular Phylogenetics and Evolution, 139, 106549. https://doi.org/10.1016/j.ympev.2019.106549
Candan, K., Kornilios, P., Ayaz, D., Kumlutaş, Y., Gül, S., Yıldırım-Caynak, E. & Ilgaz, Ç. (2021) Cryptic genetic structure within Valentin’s Lizard, Darevskia valentini (Boettger, 1892) (Squamata, Lacertidae), with implications for systematics and origins of parthenogenesis. Systematics and Biodiversity, 19 (7), 665–681. https://doi.org/10.1080/14772000.2021.1909171
Chernomor, O., von Haeseler, A. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology, 65 (6), 997–1008. https://doi.org/10.1093/sysbio/syw037
Clarke, K.R. (1988) Detecting change in benthic community structure. In: Oger, R (Ed.), Proceedings of invited papers, 14th International biometric conference. Namour, Belgium, pp. 131–142.
Clarke, K.R. (1993) Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology, 18, 117–143. https://doi.org/10.1111/j.1442-9993.1993.tb00438.x
Clement, M., Posada, D. & Crandall, K.A. (2000) TCS: A computer program to estimate gene genealogies. Molecular Ecology, 9 (10), 1657–1660. https://doi.org/10.1046/j.1365-294x.2000.01020.x
Darevsky, I.S. & Eiselt, J. (1967) Ein neuer Name für Lacerta saxicola mehelyi Lantz & Cyrén 1936. Annalen des Naturhistorischen Museums in Wien, 70, 107.
Darevsky, I.S. (1967) Rock lizards of the Caucasus: systematics, ecology and phylogenesis of the polymorphic groups of Caucasian rock lizards of the subgenus Archaeolacerta. Nauka, Leningrad. [unknown pagination]
Darevsky, I.S. (1972) Zur Verbreitung einiger Felseidechsen des Subgenus Archaeolacerta in der Türkei. Bonner Zoologische Beiträge, 23 (4), 347–351.
Darevsky, I.S. & Lukina, G.P. (1977) Rock lizards of the Lacerta saxicola Eversmann group (Sauria, Lacertidae) collected in Turkey by Richard and Erica Clark. Proceedings of the Zoological Institute of the Academy of Sciences, U.S.S.R., 1977, 60–63.
Darwin, C. (1859) On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray, London, xiv + 596 pp. https://doi.org/10.5962/bhl.title.82303
Dobzhansky, T. (1937) Genetics and the origin of species. Columbia Biological Series No. 11. Columbia University Press, New York, xvi + 364 pp.
Drummond, A.J., Suchard, M.A., Xie, D. & Rambaut, A. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973. https://doi.org/10.1093/molbev/mss075
Dufresnes, C., Strachinis, I., Suriadna, N., Mykytynets, G., Cogalniceanu, D., Székely, P., Vukov, T., Arntzen, J.W., Wielstra, B., Lymberakis, P., Geffen, E., Gafny, S., Kumlutaş, Y., Ilgaz, Ç., Candan, K., Mizsei, E., Szabolcs, M., Kolenda, K., Smirnov, N., Géniez, P., Lukanov, S., Crochet, P.A., Dubey, S., Perrin, N., Litvinchuk, S.N. & Denoël, M. (2019) Phylogeography of a cryptic speciation continuum in Eurasian spadefoot toads (Pelobates). Molecular Ecology, 28, 3257–3270. https://doi.org/10.1111/mec.15133
Durfort, M. (1978) Tècniques de transparentat d’invertebrats i d’esquelets de vertebrats: aplicacions. Circular Institució Catalana D’Història Natural, 1, 1–9.
Eiselt, J., Darevsky, I.S. & Schmidtler, J.F. (1992) Untersuchungen an Felseneidechsen (Lacerta saxicola komplex) in der östlichen Türkei, I. Lacerta valentini Boettger. Annalen des Naturhistorischen Museums in Wien, 93 (B), 1–18.
Felsenstein, J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution. International Journal of Organic Evolution, 39 (4), 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fraser, D.J. & Bernatchez, L. (2001) Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Molecular Ecology, 10, 2741–2752. https://doi.org/10.1046/j.1365-294X.2001.t01-1-01411.x
Freitas, S., Vavakou, A., Arakelyan, M., Drovetski, S.V., Crnobrnja-isailović, J., Kidov, A.A., Cogălniceanu, D., Corti, C., Lymberakis, P., Harris, D.J. & Carretero, M.A. (2016a) Cryptic diversity and unexpected evolutionary patterns in the meadow lizard, Darevskia praticola (Eversmann, 1834). Systematics and Biodiversity, 14 (2), 184–197. https://doi.org/10.1080/14772000.2015.1111267
Freitas, S., Rocha, S., Campos, J., Ahmadzadeh, F., Corti, C., Sillero, N., Ilgaz, Ç., Kumlutaş, Y., Arakelyan, M., Harris, D.J. & Carretero, M.A. (2016b) Parthenogenesis through the ice ages: A biogeographic analysis of Caucasian rock lizards (genus Darevskia). Molecular Phylogenetics and Evolution, 102, 117–127. https://doi.org/10.1016/j.ympev.2016.05.035
Gabelaia, M., Tarkhnishvili, D. & Adriaens, D. (2018) Use of three-dimensional geometric morphometrics for the identification of closely related species of Caucasian rock lizards (Lacertidae: Darevskia). Biological Journal of the Linnean Society, 125, 709–717. https://doi.org/10.1093/biolinnean/bly143
Gabelaia, M. (2019) Phylogeny and morphological variation in the rock lizards of the genus Darevskia. Thesis, Ilia State University and Ghent University, Tbilisi, 121 pp.
Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59 (3), 307–321. https://doi.org/10.1093/sysbio/syq010
Hintze, J. (2007) NCSS, PASS and GESS.Number Cruncher Statistical Systems. Kaysville, Utah. Available from: http://www.NCSS.com (accessed 31 October 2022)
Jablonski, D., Kukushkin, O.V., Avcı, A., Bunyatova, S., Kumlutaş, Y., Ilgaz, Ç., Polyakova, E., Shiryaev, K., Tuniyev, B. & Jandzik, D. (2019) The biogeography of Elaphe sauromates (Pallas, 1814), with a description of a new rat snake species. PeerJ, 7, e6944. https://doi.org/10.7717/peerj.6944
Jones, G.R. (2015) Species delimitation and phylogeny estimation under the multispecies coalescent. bioRxiv, 010199, 1–24. https://doi.org/10.1101/010199
Jones, G.R. (2017) Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology, 74 (1–2), 447–467. https://doi.org/10.1007/s00285-016-1034-0
Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L. S. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14 (6), 587–589. https://doi.org/10.1038/nmeth.4285
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A. & Flouri, T. (2017) Multi-rate Poisson tree processes for singlelocus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics, 33 (11), 1630–1638. https://doi.org/10.1101/063875
Karakasi, D., Ilgaz, Ç., Kumlutaş, Y., Candan, K., Güçlü, Ö., Kankılıç, T., Beşer, N., Sindaco, R., Lymberakis, P. & Poulakakis, N. (2021) More evidence of cryptic diversity in Anatololacerta species complex Arnold, Arribas and Carranza, 2007 (Squamata: Lacertidae) and re-evaluation of its current taxonomy. Amphibia-Reptilia, 42 (2), 201–216. https://doi.org/10.1163/15685381-bja10045
Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability, outlines version 7. Molecular Biology and Evolution, 30 (4), 772–780. https://doi.org/10.1093/molbev/mst010
Koç, H., Kutrup, B., Eroğlu, O., Bülbül, U., Kurnaz, M., Afan, F. & Eroğlu, A.İ. (2017) Phylogenetic relationships of D. rudis (Bedriaga, 1886) and D. bithynica (Mehely, 1909) based on microsatellite and mitochondrial DNA in Turkey. Mitochondrial DNA, Part A, 28 (6), 814–825. https://doi.org/10.1080/24701394.2016.1197215
Koç, H., Kutrup, B., Bülbül, U. & Kurnaz, M. (2021) The Allelic Variants in Microsatellite Loci and Phylogenetic Relationships of Darevskia rudis (Bedriaga, 1886) and D. bithynica (Méhely, 1909) Based on Mitochondrial DNA in Turkey. Russian Journal of Herpetology, 28 (2), 73–88. https://doi.org/10.30906/1026-2296-2021-28-2-73-88
Kornilios, P., Kumlutaş, Y., Lymberakis, P. & Ilgaz, Ç. (2018) Cryptic diversity and molecular systematics of the Aegean Ophiomorus skinks (Reptilia: Squamata), with the description of a new species. Journal of Zoological Systematics and Evolutionary Research, 56 (3), 364–381. https://doi.org/10.1111/jzs.12205
Kotsakiozi, P., Jablonski, D., Ilgaz, Ç., Kumlutaş, Y., Avcı, A., Meiri, S., Itescu, Y., Kukushkin, O., Gvoždík, V., Scillitani, G., Roussos, S.A., Jandzik, D., Kasapidis, P., Lymberakis, P. & Poulakakis N. (2018) Multilocus phylogeny and coalescent species delimitation in Kotschy’s gecko, Mediodactylus kotschyi: Hidden diversity and cryptic species. Molecular Phylogenetics and Evolution, 125, 177–187. https://doi.org/10.1016/j.ympev.2018.03.022
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018) MEGA X Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. https://doi.org/10.1093/molbev/msy096
Lantz, L.A. & Cyrén, O. (1936) Description of Darevskia bithynica tristis. In: Contribution à la connaissance de Lacerta saxicola Eversmann. Bulletin de la Societé Zoologique de France, Paris, 61, pp. 159–181.
Legendre, P. & Legendre, L. (1998) Numerical Ecology. Elsevier Science B. V., Amsterdam, 853 pp.
Leigh, J.W. & Bryant, D. (2015) PopART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6 (9), 1110–1116. https://doi.org/10.1111/2041-210X.12410
Mayr, E. (1982) Processes of speciation in animals. In: Liss, A.R.I. (Ed.), Mechanisms of speciation. Alan R. Liss, Inc., New York, New York, pp. 1–19.
Mendes, J., Salvi, D., Harris, D.J., Els, J. & Carranza, S. (2018) Hidden in the Arabian Mountains: Multilocus phylogeny reveals cryptic diversity in the endemic Omanosaura lizards. Journal of Zoological Systematics and Evolutionary Research, 56, 395–407. https://doi.org/10.1111/jzs.12210
Minh, B.Q., Nguyen, M.A. & von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30 (5), 1188–1195. https://doi.org/10.1093/molbev/mst024
Murphy, R.W., Fu, J., MacCulloch, R. Darevsky, I.S. & Kupriyanova, L. (2000) A fine line between sex and unisexuality: the phylogenetic constraints on parthenogenesis in lacertid lizards. Zoological Journal of the Linnean Society, 130, 527–549. https://doi.org/10.1111/j.1096-3642.2000.tb02200.x
Prohl, H., Ron, S.R. & Ryan, M.J. (2010) Ecological and genetic divergence between two lineages of Middle American tungara frogs Physalaemus (= Engystomops) pustulosus. BMC Ecology and Evolution, 10, 146. https://doi.org/10.1186/1471-2148-10-146
Rambaut, A., Drummond, A.J., Xie, D., Baele, G. & Suchard, M.A. (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67 (5), 901–904. https://doi.org/10.1093/sysbio/syy032
Rato, C., Stratakis, M., Sousa-Guedes, D., Sillero, N., Corti, C., Freitas, S., Harris, D.J. & Carretero, M.A. (2021) The more you search, the more you find: Cryptic diversity and admixture within the Anatolian rock lizards (Squamata, Darevskia). Zoologica Scripta, 50 (2), 193–209. https://doi.org/10.1111/zsc.12462
Reid, N.M. & Carstens, B.C. (2012) Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Ecology and Evolution, 12, 196. https://doi.org/10.1186/1471-2148-12-196
Rohlf, J. (2000) NTSYSpc. Version 2.1. UserGuide. Exeter Software ed., Setauket, New York, New York, 38 pp.
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.I., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61 (3), 539–542. https://doi.org/10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J.C., GuiraoRico, S., Librado, P., Ramos-Onsins, S.E. & SanchezGracia, A. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34 (12), 3299–3302. https://doi.org/10.1093/molbev/msx248
Seaby, R.M.H. & Henderson, P.A. (2019) Community Analysis Package 6.0. Pisces Conservation Ltd, Lymington, 164 pp. [http://www.pisces-conservation.com]
Sindaco, R., Kornilios, P., Sacchi, R. & Lymberakis, P. (2014) Taxonomic reassessment of Blanus strauchi (Bedriaga, 1884) (Squamata: Amphisbaenia: Blanidae), with the description of a new species from south-east Anatolia (Turkey). Zootaxa, 3795 (3), 311–326. https://doi.org/10.11646/zootaxa.3795.3.6
Sokal, R.R. & Rohlf, J. (1969) Biometry. The principles and practice of statistics in Biological research. W.F. Freeman and C., New York, New York, 776 pp.
Stephens, M., Smith, N.J. & Donnelly, P. (2001) A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics, 68 (4), 978–989. https://doi.org/10.1086/319501
Stephens, M. & Scheet, P. (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missingdata imputation. American Journal of Human Genetics, 76 (3), 449–462. https://doi.org/10.1086/428594
Şekercioğlu, Ç.H., Anderson, S., Akçay, E., Bilgin, R., Can, Ö.E., Semiz, G., Tavşanoğlu, Ç., Yökeş, M.B., Soyumert, A., İpekdal, K., Sağlam, İ.K., Yücel, M. & Dalfes, H.N. (2011) Turkey’s globally important biodiversity in crisis. Biological Conservation, 144, 2752–2769. https://doi.org/10.1016/j.biocon.2011.06.025
Tamar, K., Carranza, S., In den Bosch, H., Sindaco, R., Moravec, J. & Meiri, S. (2015) Hidden relationships and genetic diversity: Molecular phylogeny and phylogeography of the Levantine lizards of the genus Phoenicolacerta (Squamata: Lacertidae). Molecular Phylogenetics and Evolution, 91, 86–97. https://doi.org/10.1016/j.ympev.2015.05.002
Tarkhnishvili, D. (2012) Evolutionary history, habitats, diversification, and speciation in Caucasian rock lizards. Advances in Zoology Research, 2, 79–120.
Tarkhnishvili, D., Murtskhvaladze, M. & Gavashelishvili, A. (2013) Speciation in Caucasian lizards: Climatic dissimilarity of the habitats is more important than isolation time. Biological Journal of the Linnean Society, 109 (4), 876–892. https://doi.org/10.1111/bij.12092
Tarkhnishvili, D., Gabelaia, M. & Adriaens, D. (2020b) Phenotypic divergence, convergence and evolution of Caucasian rock lizards (Darevskia). Biological Journal of the Linnean Society, 130, 142–155. https://doi.org/10.1093/biolinnean/blaa021
Tarkhnishvili, D., Yanchukov, A., Şahin, M.K., Gabelaia, M., Murtskhvaladze, M., Candan, K., Galoyan, E., Arakelyan, M., Iankoshvili, G., Kumlutaş, Y., Ilgaz, Ç., Matur, F., Çolak, F., Erdolu, M., Kurdadze, S., Barateli, N. & Anderson, C. (2020a) Genotypic similarities among the parthenogenetic rock lizards Darevskia with presumed different hybrid origins. BMC Evolutionary Biology, 20, 122. https://doi.org/10.1186/s12862-020-01690-9.
Taylor, W.R. (1967) An enzyme method of clearing and staining small vertebrates. Proceedings United States National Museum, Smithsonian Institute, 122 (3596), 1–17. https://doi.org/10.5479/si.00963801.122-3596.1
Trifinopoulos, J., Nguyen, L.T., Von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44 (W1), 232–235. https://doi.org/10.1093/nar/gkw256
Uetz, P., Freed, P. & Hošek, J. (2022) The Reptile Database. Available from: http://www.reptile-database.org, (accessed 10 March 2022)
Yanchukov, A., Tarkhnishvili, D., Erdolu, M., Şahin, M.K., Candan, K., Murtskhvaladze, M., Gabelaia, M., Iankoshvili, G., Barateli, N., Ilgaz, Ç., Kumlutaş, Y., Matur, F., Çolak, F., Arakelyan, M. & Galoyan, E. (2022) Precise paternal ancestry of hybrid unisexual ZW lizards (genus Darevskia: Lacertidae: Squamata) revealed by Z-linked genomic markers. Biological Journal of the Linnean Society, 136 (2), 293–305. https://doi.org/10.1093/biolinnean/blac023
Yaşar, Ç., Çiçek, K., Mulder, J. & Tok, C.V. (2021) The distribution and biogeography of amphibians and reptiles in Turkey. North-Western Journal of Zoology, 17 (2), e201512, 232–275.
Yousefabadi, F., Rastegar-Pouyani, E., Keikhosravi, A., Rastegar Pouyani, N., Avcı, A., Üzüm, N., Olgun, K., Kumlutaş, Y., Lymberakis, P., Ilgaz, Ç. & Hosseinian Yousefkhani, S.S. (2021) An integrative approach uncovered variation within Trapelus ruderatus (Olivier, 1804) (Squamata: Agamidae) in Western Asia. Journal of Zoological Systematics and Evolutionary Research, 59, 1530–1545. https://doi.org/10.1111/jzs.12557