Abstract
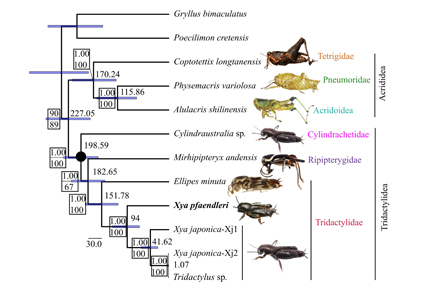
We report the comparative examination of the complete mitochondrial genome of the pygmy mole cricket Xya pfaendleri (Orthoptera: Caelifera: Tridactylidae). The mitogenome consists of 13 protein-coding regions, 22 tRNAs, two rRNAs, and one control region, following the gene order of the ancestral pancrustacean mitogenome. The length of the mitogenome in Xya pfaendleri is 15352 bp. The start and stop codons of the protein-coding genes exhibit the general pattern observed in orthopterans. The data indicate that the pattern of gene overlapping/intergenic sequences exhibits a significant phylogenetic signal. A phylogenetic tree inferred using 12 mitogenomes (seven belonging to Tridactylidea, three to Acrididea, and two to Ensifera) confirms the sister group relationship of Acrididea and Tridactylidea. The relationship among the families of Tridactylidea is Cylindrachetidae + (Ripipterygidae + Tridactylidae). The mitogenome sequences of Xya and Tridactylus constitute a single clade, sharing a last common ancestor 94 million years ago, and rendering the first genus paraphyletic. The present preliminary data suggest that we still have much to learn about the evolution and diversity of Tridactylidea.
References
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed 15 October 2023)
- Avise, J.C., Arnold, J., Ball, R.M., Bermingham, E., Lamb, T., Neigel, J.E., Reeb, C.A. & Saunders, N.C. (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics, 18 (1), 489–522. https://doi.org/10.1146/annurev.es.18.110187.002421
- Baehr, M. (1988) On Australian Tridactylidae mainly from Northern Australia (Orthopteroidea, Saltatoria, Caelifera). Spixiana, Munich, 11 (1), 13–26.
- Bernt, M., Bleidorn, C., Braband, A., Dambach, J., Donath, A., Fritzsch, G., Golombek, A. Hadrys, H., Jühling, J., Meusemann, K., Middendorf, M., Misof, B., Perseke, M., Podsiadlowski, L., Reumont, B., Schierwater, B., Schlegel, M., Schrödl, M., Simon, S., Stadler, P.F., Stöger I. & Struck, T.H. (2013) A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Molecular Phylogenetics and Evolution, 69 (2), 352–364. https://doi.org/10.1016/j.ympev.2013.05.002
- Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., Pütz, J., Middendorf, M. & Stadler, P.F. (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69 (2), 313–319.
- https://doi.org/10.1016/j.ympev.2012.08.023
- Boore, J.L. & Brown, W.M. (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Current Opinion in Genetics & Development, 8, 668–674. https://doi.org/10.1016/s0959-437x(98)80035-x
- Boore, J.L. (1999) Animal mitochondrial genomes. Nucleic Acids Research, 27 (8), 1767–1780. https://doi.org/10.1093/nar/27.8.1767
- Cameron, S. (2014a) How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Systematic Entomology, 39 (3), 400–411. https://doi.org/10.1111/syen.12071
- Cameron, S.L. (2014b) Insect mitochondrial genomics: implications for evolution and phylogeny. Annual Review of Entomology, 59, 95–117. https://doi.org/10.1146/annurev-ento-011613-162007
- Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
- Chang, H., Qiu, Z., Yuan, H., Wang, X., Li, X., Sun, H., Guo, X., Lu, Y., Feng, X., Majid, M. & Huang, Y. (2020) Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Molecular Phylogenetics and Evolution, 145, 106734. https://doi.org/10.1016/j.ympev.2020.106734
- Chikhi, R. & Rizk, G. (2013) Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithms for Molecular Biology, 8, 1–9. https://doi.org/10.1186/1748-7188-8-22
- Cigliano, M.M., Braun, H., Eades, D.C. & Otte, D. (2023) Orthoptera Species File. Version 5.0/5.0. Available from: http://orthoptera.speciesfile.org (accessed 3 July 2023)
- Fenn, J.D., Song, H., Cameron, S.L. & Whiting, M.F. (2008) A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Molecular Phylogenetics and Evolution, 49, 59–68. https://doi.org/10.1016/j.ympev.2008.07.004
- Günther, K.K. (1979) Einige Bemerkungen über die Gattungen der Familie Tridactylidae Brunner und zur Klassifi kation der Tridactylodea. Deutsche Entomologische Zeitschrift, 26, 255–264. https://doi.org/10.1002/mmnd.19790260408
- Günther, K.K. (1980) Katalog der Caelifera-Unterordnung Tridactyloidea (Insecta). Deutsche Entomologische Zeitschrift, 27, 149–178. https://doi.org/10.1002/mmnd.19800270114
- Günther, K.K. (1990) Zwei neue Xya-Arten aus dem Mittelmeergebiet (Orthoptera, Tridactylidae). Deutsche Entomologische Zeitschrift, 37, 119–136. https://doi.org/10.1002/mmnd.19900370125
- Günther, K.K., (1994) Die Tridactyloidea-Fauna Kolumbiens (Orthoptera, Caelifera). Deutsche Entomologische Zeitschrift, 41, 1–56. https://doi.org/10.1002/mmnd.19940410102
- Hassanin, A., Leger, N. & Deutsch, J. (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Systematic Biology, 54 (2), 277–298. https://doi.org/10.1080/10635150590947843
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14 (6), 587–589. https://doi.org/10.1038/nmeth.4285
- Karakaş, M.Y., Yahyaoğlu, Ö., Uluar, O., Budak, M. & Çiplak, B. (2023) Mitochondrial genome of Poecilimon cretensis (Orthoptera: Tettigoniidae: Phaneropterinae): Strong phylogenetic signals in gene overlapping regions. Zootaxa, 5263 (1), 141–147. https://doi.org/10.11646/zootaxa.5263.1.9
- Karşi, U. & Ciplak, B. (2019) Mitogenome of Anterastes babadaghi (Orthoptera: Tettigoniinae; Platycleidini): frequent conserved overlapping regions within Tettigoniinae. Zootaxa, 4651 (1), 173–190. https://doi.org/10.11646/zootaxa.4651.1.11
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20 (4), 1160–1166. https://doi.org/10.1093/bib/bbx108
- Kim, I., Cha, S.Y., Yoon, M.H., Hwang, J.S., Lee, S.M., Sohn, H.D. & Jin, B.R. (2005) The complete nucleotide sequence and gene organization of the mitochondrial genome of the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Gene, 353 (1), 155–168. https://doi.org/10.1016/j.gene.2005.04.019
- Laslett, D. & Canbäck, B. (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics, 24 (2), 172–175. https://doi.org/10.1093/bioinformatics/btm573
- Li, R. (2021) First record of the complete mitochondrial genome of Coptotettix longtanensis (Orthoptera: Tetrigidae) and its phylogenetic analysis. Available from: https://www.ncbi.nlm.nih.gov/nuccore/OK540319 (accessed 3 June 2023)
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., Von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37 (5), 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Öztürk, P.N. & Çıplak, B. (2019) Phylomitogenomics of Phaneropteridae (Orthoptera): Combined data indicate a poorly conserved mitogenome. International Journal of Biological Macromolecules, 132, 1318–1326. https://doi.org/10.1016/j.ijbiomac.2019.04.011
- Park, B., Choi, E.H., Kim, G., Shin, C.R., Hwang, J., Baek, S.Y. & Hwang, U.W. (2021) The complete mitochondrial genome of the two-spotted cricket Gryllus bimaculatus (Orthoptera: Gryllidae) from South Korea. Mitochondrial DNA Part B, 6 (3), 1144–1146. https://doi.org/10.1080/23802359.2021.1901617
- Ramesh, A., Small, S.T., Kloos, Z.A., Kazura, J.W., Nutman, T.B., Serre, D. & Zimmerman, P.A. (2012) The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Molecular and Biochemical Parasitology, 183, 32–41. https://doi.org/10.1016/j.molbiopara.2012.01.004
- Sheffield, N.C., Hiatt, K.D., Valentine, M.C., Song, H. & Whiting, M.F. (2010) Mitochondrial genomics in Orthoptera using MOSAS. Mitochondrial DNA, 21 (3–4), 87–104. https://doi.org/10.3109/19401736.2010.500812
- Song, H., Amédégnato, C., Cigliano, M.M., Desutter-Grandcolas, L., Heads, S.W., Huang, Y., Otte, D. & Whiting, M.F. (2015) 300 million years of diversification: elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics, 31 (6), 621–651. https://doi.org/10.1111/cla.12116
- Song, N., Li, H., Song, F. & Cai, W. (2016) Molecular phylogeny of Polyneoptera (Insecta) inferred from expanded mitogenomic data. Scientific Reports, 6 (1), 36175. https://doi.org/10.1038/srep36175
- Song, H., Béthoux, O., Shin, S., Donath, A., Letsch, H., Liu, S., McKenna, D.D., Meng, G., Misof, B., Podsiadlowski, L., Zhou, X., Wipfler, B. & Simon, S. (2020) Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nature communications, 11(1), 4939. https://doi.org/10.1038/s41467-020-18739-4
- Suchard, M.A., Lemey, P., Baele, G., Ayres, D.L., Drummond, A.J. & Rambaut, A. (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4 (1), vey016. https://doi.org/10.1093/ve/vey016
- Talavera, G. & Castresana, J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56, 564–577. https://doi.org/10.1080/10635150701472164
- Xu, H., Mao, B., Storozhenko, S.Y., Huang, Y., Chen, Z. & Huang, J. (2021) Phylogenetic position of the genus Alulacris (Orthoptera: Acrididae: Melanoplinae: Podismini) revealed by complete mitogenome evidence. Insects, 12 (10), 918. https://doi.org/10.3390/insects12100918
- Wang, P., Zhi, Y. & Zhang, D. (2013) The complete mitochondrial genome of Tridactylus japonicus (Orthoptera: Tridactyloidea). Available from: https://www.ncbi.nlm.nih.gov/nuccore/KC555032 (accessed 3 June 2023)