Abstract
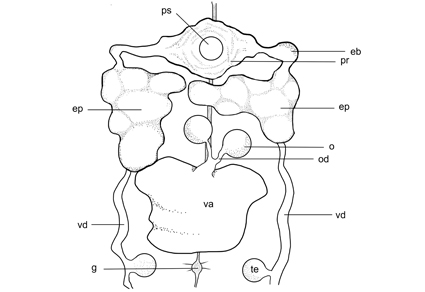
This paper presents two novel terrestrial leech species, Haemadipsa nigrilineata sp. nov. and Haemadipsa medogensis sp. nov., collected from forested regions in Tibet, China. On the basis of morphological observations, mitogenomic sequence analysis and the mitochondrial COI gene sequence phylogenetic inference, both species are herein described as new within the genus Haemadipsa Tennent, (Haemadipsidae). These two species are in line with the general diagnosis of the genus Haemadipsa in terms of living environment, annulation pattern, having five pairs of eyespots, and the position of the gonopores (XI b5/b6 and XII b5/b6). The diagnostic features for the new species include: Haemadipsa nigrilineata sp. nov. possesses a continuous jet-black longitudinal stripe along the dorsal midline and has a vaginal sac large and quadrate; Haemadipsa medogensis sp. nov. exhibits scattered irregular patterns on the dorsum and the vaginal sac is significantly large, being two to three times as long as it is wide, and the epididymis presents a dagger-like shape. Complete mitogenome sequencing revealed circular genomes of 14,755 bp for Haemadipsa nigrilineata sp. nov. and 14,892 bp for Haemadipsa medogensis sp. nov. Both genomes exhibit canonical invertebrate mitochondrial architecture, including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a non-coding control region. The phylogenetic analysis based on the mitochondrial genome showed that Haemadipsa medogensis sp. nov. is sister to H. crenata while Haemadipsa nigrilineata sp. nov. forms an independent lineage sister to H. hainana, H. yanyuanensis, Haemadipsa medogensis sp. nov. and H. crenata.
References
- Blanchard, R. (1892) Description de la Xerobdella lecomtei. Memoires de la Societe Zoologique de France, 5, 539–553.
- Borda, E., Oceguera-Figueroa, A. & Siddall, M.E. (2008) On the classification, evolution and biogeography of terrestrial haemaderoid leeches (Hirudinida: Arhynchobdellida: Hirudiniformes). Molecular Phylogenetics and Evolution, 46, 142–154. https://doi.org/10.1016/j.ympev.2007.09.006
- Borda, E. & Siddall, M.E. (2010) Insights into the evolutionary history of Indo-Pacific bloodfeeding terrestrial leeches (Hirudinida: Arhynchobdellida: Haemadipisdae). Invertebrate Systematics, 24, 456–472. https://doi.org/10.1071/IS10013
- Ding, L.M. (2004) Methods for collection, preservation and microscope-slide-preparation of bloodsucking leeches. Port Health Control, 9, 14–15. [in Chinese]
- Donath, A., Jühling, F., Al-Arab, M., Bernhart, S.H., Reinhardt, F., Stadler, P.F., Middendorf, M. & Bernt, M. (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Research, 47, 10543–10552. https://doi.org/10.1093/nar/gkz833
- Grant, J.R., Enns, E., Marinier, E., Mandal, A., Herman, E.K., Chen, C.-Y., Graham, M., Van Domselaar, G. & Stothard, P. (2023) Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Research, 51, W484–W492. https://doi.org/10.1093/nar/gkad326
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35 (2), 518–522. https://doi.org/10.1093/molbev/msx281
- Huang, T., Liu, Z., Gong, X., Wu, T., Liu, H., Deng, J., Zhang, Y., Peng, Q., Zhang, L. & Liu, Z. (2019) Vampire in the darkness: a new genus and species of land leech exclusively bloodsucking cave-dwelling bats from China (Hirudinda: Arhynchobdellida: Haemadipsidae). Zootaxa, 4560 (2), 257–272. https://doi.org/10.11646/zootaxa.4560.2.2
- Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C.W., Yi, T.-S. & Li, D.-Z. (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 21. https://doi.org/10.1186/s13059-020-02154-5
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
- Katoh, K. & Standley, D.M. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution, 30, 772–780. https://doi.org/10.1093/molbev/mst010
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P. & Drummond, A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
- Lai, Y., Nakano, T. & Chen, J. (2011) Three species of land leeches from Taiwan, Haemadipsa rjukjuana comb. n., a new record for Haemadipsa picta Moore, and an updated description of Tritetrabdella taiwana (Oka). ZooKeys, 139, 1–22. https://doi.org/10.3897/zookeys.139.1711
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution, 37, 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Magalhães, F.W., Hutchings, P., Oceguera-Figueroa, A., Martin, P., Schmelz, R.M., Wetzel, M.J., Wiklund, H., Maciolek, N.J., Kawauchi, G.Y. & Williams, J.D. (2021) Segmented worms (Phylum Annelida): a celebration of twenty years of progress through Zootaxa and call for action on the taxonomic work that remains. Zootaxa, 4979 (1), 190–211. https://doi.org/10.11646/zootaxa.4979.1.18
- Oka, A. (1910) Synopsis der jepanischen Hirudineen, mit diagnosen der neuen speices. Annotationes Zoologicae Japonenses, 7, 165–183.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
- Sawyer, R.T. (1986) Leech Biology and Behaviour. Clarendon Press and Oxford University Press, Oxford, 1065 pp.
- Singla, D. & Yadav, I.S. (2022) GAAP: A GUI-based Genome Assembly and Annotation Package. Current Genomics, 23, 77–82. https://doi.org/10.2174/1389202923666220128155537
- Song, D. (1977) Research on Several Species of Blood-Sucking Leeches in China. Current Zoology, 01, 102–108.
- Struck, T.H., Golombek, A., Hoesel, C., Dimitrov, D. & Elgetany, A.H. (2023) Mitochondrial Genome Evolution in Annelida—A Systematic Study on Conservative and Variable Gene Orders and the Factors Influencing its Evolution. Systematic Biology, 72, 925–945. https://doi.org/10.1093/sysbio/syad023
- Tan, E.-G. (2008) Progress in the study of ecology, zoogeography, group, control repellent and medical usage of Hirudinea in China. Acta Ecologica Sinica, 28, 6272–6281. https://doi.org/10.1016/S1872-2032(09)60016-0
- Tan, E.-G., Pan, X.-G. & Feng, Q.-Y. (2000) A new subspecies of Haemadipsa guangchuanensis chuandianensis from Sichuan Province, China (Haemadipsidae). Natural Science Journal of Hainan University, 18, 46–49.
- Whitman, C.O. (1886) The Leeches of Japan. Quarterly Journal of Microscopy Science, 26, 317–416. https://doi.org/10.1242/jcs.s2-26.103.317
- Yang, T. (1996) The fauna of Hirudinea in China. Science press, Beijing, 259 pp. [pp. 2–35 + 148–172, in Chinese]