Abstract
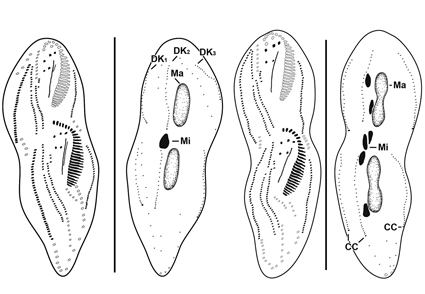
Strongylidium koreanum n. sp., a new soil ciliate from Jeju Island, South Korea, is described based on live observations, protargol impregnation, and molecular analysis of the 18S rRNA gene sequence. It is characterized by the following morphological features: cell outline more or less fusiform, posterior end broader than anterior end; grayish under low magnification; cortical granules absent; 23–32 adoral membranelles; three enlarged frontal cirri; buccal cirrus and postoral ventral cirrus present; 27–42 left and 15–28 right ventral cirri; 23–36 left and 30–46 right marginal cirri; three dorsal kineties; three caudal cirri; and two macronuclear nodules with two or three micronuclei. Phylogenetic analyses show that Strongylidium is monophyletic.
References
Chen, X., Miao, M., Ma, H., Shao, C. & Al-Rasheid, K.A. (2013) Morphology, morphogenesis and small-subunit rRNA gene sequence of the novel brackish-water ciliate Strongylidium orientale sp. nov. (Ciliophora, Hypotrichia). International journal of systematic and evolutionary microbiology, 63, 1155–1164. https://doi.org/10.1099/ijs.0.048157-0
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature methods, 9 (8), 772. https://doi.org/10.1038/nmeth.2109
Foissner, W. (2014) An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. International journal of systematic and evolutionary microbiology, 64 (1), 271–292. https://doi.org/10.1099/ijs.0.057893-0
Foissner, W., Agatha, S. & Berger, H. (2002) Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib desert. Part I: Text and line drawings. Part II: Photographs. Denisia, 5, 1–1459.
Gelei, J. (1954) Über die Lebensgemeinschaft einiger temporärer Tümpel auf einer Bergwiese im Börzsönygebirge (Oberungarn) III. Ciliaten. Acta Biologica Hungarica, 5, 259–343.
Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic biology, 59, 307–321. https://doi.org/10.1093/sysbio/syq010
Hall, T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G. & Gibson, T.J. (1998) Multiple sequence alignment with Clustal X. Trends in biochemical sciences, 23, 403–405. https://doi.org/10.1016/S0968-0004(98)01285-7
Jung, J.H., Baek, Y.S. & Min, G.S. (2011) New record of two Apokeronopsis species (Ciliophora: Urostylida: Pseudokeronopsidae) from Korea. The Korean Society of Systematic Zoology, 27 (2), 115–122. https://doi.org/10.5635/KJSZ.2011.27.2.115
Kahl, A. (1932) Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 3. Spirotricha. Die Tierwelt Deutshlands und der Angrenzenden Meeresteile, 25, 399–650.
Luo, X., Yan, Y., Shao, C., Al-Farraj, S.A., Bourland, W.A. & Song, W. (2018) Morphological, ontogenetic and molecular data support strongylidiids as being closely related to Dorsomarginalia (Protozoa, Ciliophora) and reactivation of the family Strongylidiidae Fauré-Fremiet, 1961. Zoological Journal of the Linnean Society, 184 (2), 237–254. https://doi.org/10.1093/zoolinnean/zly001
Medlin, L., Elwood, H.J., Stickel, S. & Sogin, M.L. (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71 (2), 491–499. https://doi.org/10.1016/0378-1119(88)90066-2
Paiva, T.S. 2020. Systematic redefinition of the Hypotricha (Alveolata, Ciliophora) based on combined analyses of morphological and molecular characters. Protist, 171 (4), 125755. https://doi.org/10.1016/j.protis.2020.125755
Paiva, T.S. & Silva-Neto, I.D. (2007) Morphology and morphogenesis of Strongylidium pseudocrassum Wang and Nie, 1935, with redefinition of Strongylidium Sterki, 1878 (Protista: Ciliophora: Stichotrichia). Zootaxa, 1559 (1), 31–57. https://doi.org/10.11646/zootaxa.1559.1.2
Ronquist, F. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology, 61 (3), 539–542. https://doi.org/10.1093/sysbio/sys058
Sonnenberg, R., Nolte, A.W. & Tautz, D. (2007) An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Frontiers in zoology, 4 (1), 1–12. https://doi.org/10.1186/1742-9994-4-6
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution, 28 (10), 2731–2739. https://doi.org/10.1093/molbev/msr121