Abstract
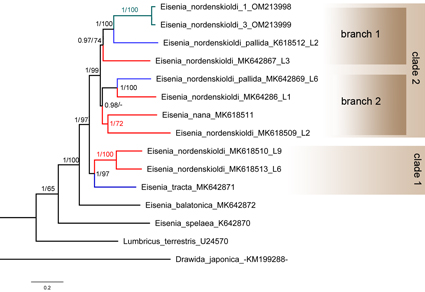
The Eisenia nordenskioldi (Eisen, 1879) species complex is one of the most widely distributed earthworm taxa in northern Asia, owing to its high morphological and genetic variability. This complex is represented by smaller, slightly pigmented- or unpigmented specimens in the Korean Peninsula. Here, we report the mitogenomes of two Eisenia nordenskioldi cf. pallida specimens, No. 1 and No. 3, which are slightly pigmented and unpigmented, respectively. The total length of the mitogenomes was 15,824 bp and 15,177 bp, respectively, and contained 37 genes, comprising 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a putative non-coding region, as in other earthworms. Nucleotide sequence comparisons of the 37 genes and the predicted protein sequences of 13 PCGs showed 96.6 % and 98.6 % similarity between the two mitogenomes, respectively. A phylogenetic analysis of 33 Crassiclitellata mitogenomes corroborated the monophyly of the Eisenia genus and recovered two highly supported subclades in the E. nordenskioldi species complex. The Korean specimens were the closest to the E. n. pallida L2 specimens collected near the type locality and probably represent the genuine E. nordenskioldi pallida Malevich, 1956.
References
Akaike, H. (1973) Information theory and an extension of the maximum likelihood principle. In: Petrow, B.N. & Csáki, F. (Eds.), Proceedings of the 2nd International Symposium on Information. Akademiai Kiado, Budapest, pp. 267–281.
Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Blakemore, R.J. (2013) Earthworms newly from Mongolia (Oligochaeta, Lumbricidae, Eisenia). ZooKeys, 285, 1–21. https://doi.org/10.3897/zookeys.285.4502
Bolger, A.M., Lohse, M. & Usadel, B. (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30 (15), 2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Boore, J.L. & Brown, W.M. (1995) Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics, 138, 423–433. https://doi.org/10.1093/genetics/141.1.305
Chang, C.-H. & James, S.W. (2011) A critique of earthworm molecular phylogenetics. Pedobiologia, 54S, 3–9. https://doi.org/10.1016/j.pedobi.2011.07.015
Decaëns T., Porco D., Rougerie R., Brown G.G. & James S.W. (2013) Potential of DNA barcoding for earthworm research in taxonomy and ecology. Applied Soil Ecology, 65, 35–42. https://doi.org/10.1016/j.apsoil.2013.01.001
Domínguez, J., Aira, M., Breinholt, J.W., Stojanovic, M., James, S.W. & Pérez-Losada, M. (2015) Underground evolution: New roots for the old tree of lumbricid earthworms. Molecular Phylogenetics and Evolution, 83, 7–19. https://doi.org/10.1016/j.ympev.2014.10.024
Donath, A., Jühling, F., Al-Arab, M., Bernhart, S.H., Reinhardt, F., Stadler, P.F., Middendorf, M. & Bernt, M. (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Research, 47(20), 10543–10552. https://doi.org/10.1093/nar/gkz833
Eisen, G. (1879) On the Oligochaeta collected during the Swedish Expeditions to the Arctic regions in the years 1870, 1875 and 1876. Öfversigt af Kunglinga Svenska Vetenskaps-Akademiens Forhandligar, 15 (7), 1–49.
Hammer, Ø., Harper, D.A.T. & Ryan, P.D. (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica, 4, 1–9.
Hong, Y. & Csuzdi, Cs. (2016) New Data to the Earthworm Fauna of the Korean Peninsula with Redescription of Eisenia koreana (Zicsi) and Remarks on the Eisenia nordenskioldi Species Group (Oligochaeta, Lumbricidae). Zoological Studies, 55 (12), 1–15. https://doi.org/10.6620/zs.2016.55-12
Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14 (6), 587–589. https://doi.org/10.1038/nmeth.4285
Kobayashi, S. (1940) Terrestrial Oligochaeta from Manchoukuo. The Science reports of the Tohoku Imperial University, 15, 261–256.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35 (6), 1547–1549. https://doi.org/10.1093/molbev/msy096
Lemoine, F., Correia, D., Lefort, V., Doppelt-Azeroual, O., Mareuil, F., Cohen-Boulakia, S. & Gascuel, O. (2019) NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Research, Web Server issue W261. https://doi.org/10.1093/nar/gkz303
Malevich, I.I. (1956) To the earthworm fauna of the Far East. Proceedings of V.P. Potemkin Moscow Municipal Pedagogical Institute, 61, 439–449. [in Russian]
Michaelsen, W. (1910) Zur Kenntnis der Lumbriciden und ihrer Verbreitung. Annuaire du Musée Zoologique de l'Académie Impériale des Sciences, St.-Petersburg, 15, 1–74
Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32 (1), 268–74. https://doi/org/10.1093/molbev/msu300.
Perel, T.S. (1969) Die Regenwürmer (Lumbricidae) des Gebirgigen Mittelasien. Pedobiologia, 9, 55–68.
Perel, T.S. (1985) Specific Features of the Earthworm Fauna (Oligochaeta, Lumbricidae) in Altai Refugia of Nemoral Vegetation. Doklady Akademii Nauk SSSR, 283 (3), 752–756. [In Russian]
Perel, T.S. (1987) The nature of eurytopy in polyploid earthworm species in relation to their use in biological amelioration. Biology and Fertility of Soils, 3: 103–105. https://doi.org/10.1007/BF00260588
Perel, T.S. (1997) The earthworms of the fauna of Russia. Cadaster and key. Nauka, Moscow, 102 pp. [In Russian]
Perel, T.S. & Graphodatsky, A.S. (1984) New species of the genus Eisenia (Lumbricidae, Oligochaeta) and their chromosome sets. Zoologicheskij Zhurnal, 63, 610–612.
Rambaut, A. (2018) FigTree version 1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 12 Jan 2022)
Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under 418 mixed models. Bioinformatics, 19 (12), 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Schwarz, G. (1978) Estimating the dimension of a model. The annals of statistics, 6 (2), 461–464. http://doi.org/10.1214/AOS/1176344136
Shekhovtsov, S.V., Golovanova, E.V. & Peltek, S.E. (2013) Cryptic diversity within the Nordenskiold’s earthworm, Eisenia nordenskioldi subsp. nordenskioldi (Lumbricidae, Annelida). European Journal of Soil Biology, 58, 13–18. https://doi.org/10.1016/j.ejsobi.2013.05.004
Shekhovtsov, S.V., Berman, D.I., Bazarova, N.E., Bulakhova, N.A., Porco, D. & Peltek, S. E. (2016) Cryptic genetic lineages in Eisenia nordenskioldi pallida (Oligochaeta, Lumbricidae). European Journal of Soil Biology, 75, 151–156. https://doi.org/10.1016/j.ejsobi.2016.06.004
Shekhovtsov, S. & Peltek, S.E. (2019) The complete mitochondrial genome of Aporrectodea rosea (Annelida: Lumbricidae). Mitochondrial DNA Part B, 4 (1), 1752–1753. https://doi.org/10.1080/23802359.2019.1610091
Shekhovtsov, S.V., Golovanova, E.V., Ershov, N.I., Poluboyarova, T.V., Berman, D.I., Bulakhova, N.A., Szederjesi, T. & Peltek, S.E. (2020a) Phylogeny of the Eisenia nordenskioldi complex based on mitochondrial genomes. European Journal of Soil Biology, 96, 103137. https://doi.org/10.1016/j.ejsobi.2019.103137
Shekhovtsov, S.V., Shipova, A.A., Poluboyarova, T.V., Vasiliev, G.V., Golovanova, E.V., Geraskina, A.P., Bulakhova, N.A., Szederjesi, T. & Peltek, S.E. (2020b) Species Delimitation of the Eisenia nordenskioldi Complex (Oligochaeta, Lumbricidae) Using Transcriptomic Data. Frontiers in Genetics, 11, 598196. https://doi.org/10.3389/fgene.2020.598196
Zhang, Q., Liu, H., Zhang, Y. & Ruan, H. (2019) The complete mitochondrial genome of Lumbricus rubellus (Oligochaeta, Lumbricidae) and its phylogenetic analysis. Mitochondrial DNA B, 4 (2), 2677–2678. https://doi.org/10.1080/23802359.2019.1644242