Abstract
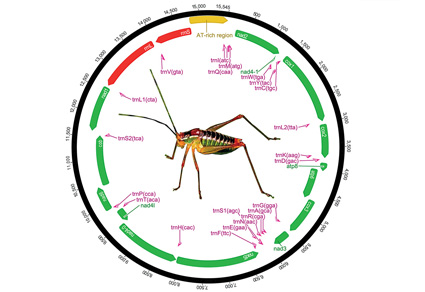
We report the complete mitochondrial genome of the Cretan bush cricket Poecilimon cretensis. The mitogenome consists of 13 protein-coding regions, 22 tRNAs, two rRNAs, and one control region. The length of mitogenome in P. cretensis varies between15477 and 15631 bp, mainly due to variability in control region. The start and stop codons of protein coding genes exhibit the general pattern in Phaneropterinae. Phylogenetic tree constructed with the mitogenome obtained during this study and 12 mitogenomes of Phaneropterinae downloaded from GenBank, placed P. cretensis in Barbitistini as sister group to Poecilimon luschani. Data indicate that the gene overlapping pattern exhibit strong phylogenetic signals.
References
- Cameron, S.L. (2014) Insect mitochondrial genomics: implications for evolution and phylogeny. Annual Review of Entomology, 59, 95–117. https://doi.org/10.1146/annurev-ento-011613-162007 DOI: https://doi.org/10.1146/annurev-ento-011613-162007
- Cıgliano, M.M., Braun, H., Eades, D.C. & Otte, D. (2021) Orthoptera Species File. Version 5.0/5.0. Available from: http://orthoptera.speciesfile.org (Accessed 25 September 2022)
- Çıplak, B. (2004) Biogeography of Anatolia: the marker group Orthoptera. Memorie Della Societa Entomologica Italiana, 82 (2), 357–372.
- Çıplak, B. (2021) Locust and grasshopper outbreaks in the Near East: Review under global warming context. Agronomy, 11, 111. https://doi.org/10.3390/agronomy11010111 DOI: https://doi.org/10.3390/agronomy11010111
- Donath, A., Jühling, F., Al-Arab, M., Bernhart, S.H., Reinhardt, F., Stadler, P.F. & Bernt M. (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Research, 47 (20), 10543–10552. https://doi.org/10.1093/nar/gkz833 DOI: https://doi.org/10.1093/nar/gkz833
- Fenn, J.D., Song, H., Cameron, S.L. & Whiting, M.F. (2008) A preliminary mitochondrial genome phylogeny of Orhoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Molecular Phylogenetics and Evolution, 49, 59–68. https://doi.org/10.1016/j.ympev.2008.07.004 DOI: https://doi.org/10.1016/j.ympev.2008.07.004
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14 (6), 587–589. https://doi.org/10.1038/nmeth.4285 DOI: https://doi.org/10.1038/nmeth.4285
- Kim, I., Cha, S.Y., Yoon, M.H., Hwang, J.S., Lee, S.M., Sohn, H.D. & Jin, B.R. (2005) The complete nucleotide sequence and gene organization of the mitochondrial genome of the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Gene, 353 (1), 155–168. https://doi.org/10.1016/j.gene.2005.04.019 DOI: https://doi.org/10.1016/j.gene.2005.04.019
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35 (6), 1547. https://doi.org/10.1093/molbev/msy096 DOI: https://doi.org/10.1093/molbev/msy096
- Laslett, D. & Canbäck, B. (2008) ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics, 24, 172–175. https://doi.org/10.1093/bioinformatics/btm573 DOI: https://doi.org/10.1093/bioinformatics/btm573
- Liu, C., Chang, J., Ma, C., Li, L. & Zhou, S. (2013) Mitochondrial genomes of two Sinochlora species (Orhoptera): novel genome rearrangements and recognition sequence of replication origin. BMC Genomics, 14, 1471–2164. https://doi.org/10.1186/1471-2164-14-114 DOI: https://doi.org/10.1186/1471-2164-14-114
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37 (5), 1530–1534. https://doi.org/10.1093/molbev/msaa015 DOI: https://doi.org/10.1093/molbev/msaa015
- Öztürk, P.N. & Çıplak, B. (2019) Phylomitogenomics of Phaneropteridae (Orthoptera): Combined data indicate a poorly conserved mitogenome. International Journal of Biological Macromolecules, 132, 1318–1326. https://doi.org/10.1016/j.ijbiomac.2019.04.011 DOI: https://doi.org/10.1016/j.ijbiomac.2019.04.011
- Tang, M., Tan, M., Meng, G. & Zhou, X. (2014) Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Research, 42 (22), e166-e166. https://doi.org/10.1093/nar/gku917 DOI: https://doi.org/10.1093/nar/gku917
- Salikhov, K., Sacomoto, G. & Kucherov, G. (2014) Using cascading bloom filters to improve the memory usage for de Brujin graphs. Algorithms for Molecular Biology, 9, 2. https://doi.org/10.1186/1748-7188-9-2 DOI: https://doi.org/10.1186/1748-7188-9-2
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics, 27 (2), 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x DOI: https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Wang, J., Qiu, Z., Yuan, H. & Huang, Y. (2017) The complete mitochondrial genomes of two Phaneroptera species (Orthoptera: Tettigoniidea) and comparative analysis of mitochondrial genomes in Orthoptera. Available from: https://www.ncbi.nlm.nih.gov/nuccore (Accessed 18 December 2018)
- Yang, J., Ye, F. & Huang, Y. (2016) Mitochondrial genomes of four katydids (Orthoptera: Phaneropteridae): new gene rearrangements and their phylogenetic implications. Gene, 702–711. https://doi.org/10.1016/j.gene.2015.09.052 DOI: https://doi.org/10.1016/j.gene.2015.09.052
- Zhou, Z., Yang, M., Chang, Y. & Shi, F. (2013) Comparative analysis of mitochondrial genomes of two long-legged katydids (Orthoptera: Tettigoniidae). Acta Entomologica Sinica, 56 (4), 408–418.
- Zhou, Z., Zhao, L., Liu, N., Guo, H., Guan, B., Di, J. & Shi, F. (2017) Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences, Molecular Phylogenetics and Evolution, 108, 22–33. https://doi.org/10.1016/j.ympev.2017.01.014 DOI: https://doi.org/10.1016/j.ympev.2017.01.014