Abstract
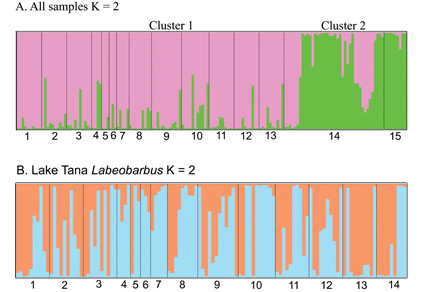
The Lake Tana Labeobarbus species flock represents one of the world’s most famous examples of lacustrine species radiations. Previous studies of this group have resulted in the description of at least 15 species based on their differences in functional morphology and definition of two clades (lacustrine and riverine spawning clades) based on life history traits. A total of 166 fish representing 14 Labeobarbus species were genotyped using 10 lineage-specific hexaploid microsatellite loci. Six of these loci were developed for this study based on DNA sequence contigs derived from a microsatellite-enriched genomic library of Labeobarbus intermedius from Lake Tana; the remaining four loci were obtained from a previous study. The genotypes of the 10 loci were analyzed to examine genetic diversity and population structure within Lake Tana Labeobarbus. Overall mean allelic richness (NA) was 17.6 alleles per locus and observed (Ho) and expected (He) heterozygosities were 0.84 ± 0.14 and 0.73 ± 0.09, respectively, across all Lake Tana Labeobarbus samples examined. Our analyses reveal that there is little genetic differentiation among species (FST = 0.020–0.099; only 10 of 91 species comparisons were significant), but moderate differentiation (FST = 0.11, p < 0.05) between lacustrine and riverine spawning populations. Relative to previous phylogenetic hypotheses, our phenetic analysis employing the R-based Analysis of Phylogenetics and Evolution (APE) program seems to perform marginally better in revealing lineages within Lake Tana Labeobarbus. Herein, our results are compared to a previous microsatellite-based study of the same populations.
References
- Albertson, R.C., Markert, J.A. Danley, P.D. & Kocher, T.D. (1999) Phylogeny of a rapidly evolving clade: the cichlid fishes of Lake Malawi, East Africa. Proceedings of the National Academy of Sciences of the USA, 96, 5107–5110. https://doi.org/10.1073/pnas.96.9.5107
- Alekseyev, S.S., Dgebuadze, Y.Y., Mina, M.V. & Mironovsky, A.N. (1996) Small ‘large barbs’ spawning in tributaries of Lake Tana: what are they? Folia Zoologica, 45, 85–96.
- Amos, W., Hoffman, J.I., Frodsham, A., Zhang, L., Best, S. & Hill, A.V.S. (2007) Automated binning of microsatellite alleles: problems and solutions. Molecular Ecology Notes, 7, 10–14. https://doi.org/10.1111/j.1471-8286.2006.01560.x
- Anteneh, W., Getahun, A., Dejen, E., Sibbing, F.A., Nagelkerke, L.A.J., de Graaf, M., Wudneh, T., Vijverberg, J. & Palstra, A.P. (2012) Spawning migrations of the endemic Labeobarbus (Cyprinidae, Teleostei) species of Lake Tana, Ethiopia: status and threats. Journal of Fish Biology, 81, 750–765. https://doi.org/10.1111/j.1095-8649.2012.03362.x
- Banister, K.E. (1973) A revision of the large Barbus (Pisces, Cyprinidae) of East and Central Africa. Studies on African Cyprinidae. Bulletin of the British Museum of Natural History (Zoology), 26, 167–180. https://doi.org/10.5962/bhl.part.204
- Berrebi, P. & Valiushok, D. (1998) Genetic divergence among morphotypes of Lake Tana (Ethiopia) barbs. Biological Journal of Linnean Society, 64, 369–384. https://doi.org/10.1111/j.1095-8312.1998.tb00338.x
- Beshera, K.A. & Harris, P.M. (2014) Mitochondrial DNA phylogeography of Labeobarbus intermedius (Cyprinidae; Pisces) from Ethiopia. Journal of Fish Biology, 85:228–245. https://doi.org/10.1111/jfb.12408
- Bini, G. (1940) I Pesci de lago Tana. Missione di Studio al Lago Tana, 3, 138–246.
- Boulenger, G.A. (1902) Descriptions of new fishes from the collection made by Mr. E. Degen in Abyssinia. Annals and Magazine of Natural History, 10, 421–437. https://doi.org/10.1080/00222930208678700
- Brown, K.M., Baltazar, G.A. & Hamilton, M.B. (2005) Reconciling nuclear microsatellite and mitochondrial marker estimates of population structure: breeding population structure of Chesapeake Bay striped bass (Morone saxatilis). Heredity, 94, 606–615. https://doi.org/10.1038/sj.hdy.6800668
- Chenuil, A., Desmarais, E., Pouyaud, L. & Berrebi, P. (1997) Does polyploidy lead to fewer and shorter microsatellites in Barbus (Teleostei: Cyprinidae)? Molecular Ecology, 6, 169–178. https://doi.org/10.1046/j.1365-294x.1997.00170.x
- Clark, L.V. & Jasieniuk, M. (2011) Polysat: An R package for polyploid microsatellite analysis. Molecular Ecology Resources, 11, 562–566. https://doi.org/10.1111/j.1755-0998.2011.02985.x
- De Graaf, M., Dejen, E., Osse, W.J.M. & Sibbing, F.A. (2008) Adaptive radiation of Lake Tana’s Labeobarbus species-flock (Pisces, Cyprinidae). Marine and Freshwater Research, 59, 391–407. https://doi.org/10.1071/MF07123
- De Graaf, M., Megens, H.J., Samallo, J. & Sibbing, F.A. (2010) Preliminary insight into the age and origin of the Labeobarbus species-flock from Lake Tana (Ethiopia) using the mtDNA cytochrome b gene. Molecular Phylogenetics and Evolution, 55, 488–500. https://doi.org/10.1016/j.ympev.2009.10.029
- De Silva, H.N., Hall, A.J., Rikkerink, M.A., McNeilage, M.A. & Fraser, L.G. (2005) Estimation of allele frequencies in polyploids under certain patterns of inheritance. Heredity, 95, 327–334. https://doi.org/10.1038/sj.hdy.6800728
- Dgebuadze, Y.Y. (1999) Observations on reproduction of Lake Tana barbs. Journal of Fish Biology, 54, 417–423. https://doi.org/10.1111/j.1095-8649.1999.tb00840.x
- Dixon, B., Nagelkerke, L.A.J., Sibbing, F.A., Egberts, E. & Stet, R.J.M. (1996) Evolution of MHC class II βchain-encoding genes in the Lake Tana barbel species-flock (Barbus intermedius complex). Immunogenetics, 44, 419–431. https://doi.org/10.1007/s002510050148 https://doi.org/10.1007/BF02602803
- Drummond, A., Ashton, B., Cheung, M., Heled, J., Kearse, M., Moir, R., Stones-Havas, S., Thierer, T. & Wilson, A. (2009) Geneious v4.7. Available from: http://www.geneious.com/ (last accessed 02 Feburary 2022)
- Dufresne, F., Stift, M., Vergilino, R. & Mable, B.K. (2014) Recent progress and challenges in population genetics of polyploid organisms: an overview of current state–of–the–art molecular and statistical tools. Molecular Ecology, 23, 40–69. https://doi.org/10.1111/mec.12581
- Dzerzhinskii, F.N., Shkil, F.N., Abdissa, B., Zelalem, W. & Mina, M.V. (2007) Spawning of Large Barbus (Barbus intermedius Complex) in a Small River of the Lake Tana Basin (Ethiopia) and Relationships of Some Putative Species. Journal of Ichthyology, 47, 639–646. https://doi.org/10.1134/S0032945207080103
- Edwards, D.D., Vidrine, M.F. & Ernsting, B.R. (2010) Phylogenetic relationships among Unionicola (Acari: Unionicolidae) mussel–mites of North America based on mitochondrial cytochrome c oxidase I sequences. Zootaxa, 2537 (1), 47–50. https://doi.org/10.11646/zootaxa.2537.1.4
- Evanno, G., Regnaut, S. & Goudet, J. (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology, 14, 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
- Golubtsov, A.S. & Krysanov, E.Y. (1993) Karyological study of some cyprinid species from Ethiopia. The ploidy differences between large and small Barbus of Africa. Journal of Fish Biology, 42, 445–455. https://doi.org/10.1111/j.1095-8649.1993.tb00347.x
- Guégan, J.-F, Rab, P., Machordom, A. & Doadrio, I. (1995) New evidence of hexaploidy in 'large' African Barbus with some considerations on the origin of hexaploidy. Journal of Fish Biology, 4, 192–198. https://doi.org/10.1111/j.1095-8649.1995.tb01888.x
- Hamrick, J.L., Godt, M.J.W., Murawshi, D.A. & Loveless, M.D. (1991) Correlations between species traits and allozyme diversity: implications for conservation biology. In: Falk, D.A. & Holsinger, K.E. (Eds.), Genetics and conservation of rare plants. Oxford University Press, New York, pp. 75–86.
- Hillis, D.M., Mable, B.K., Larson, A., Davis, S.K. & Zimmer, E.A. (1996) Nucleic acids IV: sequencing and cloning. In: Hillis, D., Moritz, C. & Mable, B. (Eds.), Molecular Systematics. Sinauer Associates, Sunderland, MA.
- Jombart, T., Devillard, S. & Balloux, F. (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics, 11, 9. https://doi.org/10.1186/1471-2156-11-94
- Kruiswijk, C.P., Hermsen, T., Heerwaarden, J.V., Dixon, B., Savelkoul, H.F.J. & Stet, R.J.M. (2005) Major histocompatibility genes in Lake Tana African large barb species-flock; evidence for complete partitioning of class II B, but not class I, genes among different species. Immunogenetics, 56, 894–908. https://doi.org/10.1007/s00251-005-0767-5. Epub 2005 Feb 8
- Krysanov, E., Yu, E. & Golubtsov, A.S. (1996) Karyotypes of some Ethiopian Barbus and Varicorhinus from the Nile Basin including Lake Tana morphotypes. Folia Zoologica, 45 (1), 67–75.
- Levin, B.A., Casal-López, M., Simonov, E., Dgebuadze, Y.Y., Mugue, N.S., Tiunov, A.V., Doadrio, I. & Golubstov, A.S. (2019) Adaptive radiation of barbs of the genus Labeobarbus (Cyprinidae) in the East African river. Freshwater Biology, 64, 1721–1736. https://doi.org/10.1111/fwb.13364
- Levin, B.A., Dgebuadze, Y.Y., Tefera, F., Tesfaye, G. & Golubstov, A.S. (2017) An evidence of past introgressive hybridization between Labeobarbus ethiopicus and Labeobarbus intermedius in the Ethiopian rift Valley, East Africa. Ethiopian Journal of Biological Sciences, 16 (Suppl.), 45-60.
- Levin, B.A., Komarova, A.S., Rozanova, O.L. & Golubtsov, A.S. (2021) Unexpected Diversity of Feeding Modes among Chisel–Mouthed Ethiopian Labeobarbus (Cyprinidae). Water, 13, 2345. https://doi.org/10.3390/w13172345
- Levin, B.A., Simonov, E., Dgebuadze, Y.Y., Levina, M. & Golubstov, A.S. (2020) In the rivers: multiple adaptive radiations of cyprinid fishes (Labeobarbus) in Ethiopian highlands. Scientific Reports, 10, 7192. https://doi.org/10.1038/s41598-020-64350-4
- Levin, B.A., Simonov, E., Franchini, P., Mugue, N., Golubtsov, A. & Meyer, A. (2021). Rapid adaptive radiation in a hillstream cyprinid fish in the East African White Nile River basin. Molecular Ecology, 30 (21), 5530–5550. https://doi.org/10.1111/mec.16130.
- Levin, B., Simonov, E., Gabrielyan, B.K., Mayden, R.L., Rastorguev, S.M., Roubenyan, H.R., Sharko, F.S. & Nedoluzhko, A.V. (2022) Caucasian treasure: Genomics sheds light on the evolution of half-extinct Sevan trout, Salmo ischchan, species flock. Molecular Phylogenetics and Evolution, 167, 107346. https://doi.org/10.1016/j.ympev.2021.107346
- Mallet, J. (2005) Hybridization as an invasion of the genome. Trends in Ecology and Evolution, 20 (5), 229–239. https://doi.org/10.1016/j.tree.2005.02.010
- Mayer, C. (2006–2010) Phobos 3.3.11. Availabe from: http://www.rub.de/spezzoo/cm/cm_phobos.htm (Accessed 05 May 2014)
- Mayr, E. (1963) Animal species and evolution. Belknap Press, Cambridge, MA, 797 pp. https://doi.org/10.4159/harvard.9780674865327
- Meriams, P.G. & Tienderen, P.H.V. (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes, 4, 792–794. https://doi.org/10.1111/j.1471-8286.2004.00770.x
- Moody, M.E., Muellert, L.D. & Soltis, D.E. (1993) Genetic Variation and Random Drift in Autotetraploid Population. Genetics, 154, 649–657. https://doi.org/10.1093/genetics/134.2.649
- Nagelkerke, L.A.J. & Sibbing, F.A. (1996) Reproductive segregation among the large barbs (Barbus intermedius complex) of Lake Tana, Ethiopia. An example of lacustrine speciation? Journal of Fish Biology, 49, 1244–1266. https://doi.org/10.1111/j.1095-8649.1996.tb01793.x
- Nagelkerke, L.A.J. & Sibbing, F.A. (1997) A revision of the large barbs (Barbus spp., Cyprinidae, Teleostei) of Lake Tana, Ethiopia, with a description of seven new species. In: Nagelkerke, L.A.J (Ed.), The barbs of Lake Tana, Ethiopia: morphological diversity and its implications for taxonomy, trophic resource partitioning, and fisheries (Doctoral dissertation). Wageningen Agricultural University, Wageningen, pp. 105–171.
- Nagelkerke, L.A.J. & Sibbing, F.A. (2000) The large barbs (Barbus spp., Cyprinidae, Teleostei) of Lake Tana (Ethiopia), with a description of a new species, Barbus osseensis. Netherlands Journal of Zoology, 50, 179–214. https://doi.org/10.1163/156854200505946
- Nagelkerke, L.A.J., Sibbing, F.A., Boogaart, J.G.M., Lammens, E.H.R.R. & Osse, J.W.M. (1994) The barbs (Barbus spp.) of Lake Tana: a forgotten species flock? Environmental Biology of Fishes, 39, 1–22. https://doi.org/10.1007/BF00004751
- Nagelkerke, L.A.J., Leon-Kloosterziel, K.M., Megens, H.-J., de Graaf, M., Diekmann, O.E. & Sibbing, F.A. (2015) Shallow genetic divergence and species delineations in the endemic Labeobarbus species flock of Lake Tana, Ethiopia. Journal of Fish Biology, 87, 1191–208. https://doi.org/10.1111/jfb.12779
- Naran, D., Skelton, P.H. & Villet, M.H. (2007) Karyology of three evolutionarily hexaploid southern African species of yellowfish, Labeobarbus Rüppel, 1836 (Cyprinidae). African Zoology, 42, 254–260. https://doi.org/10.3377/1562-020(2007)42[254:KOTEHS]2.0.CO;2
- Nater, A., Burri, R., Kawakami, T., Smeds, L. & Ellegren, H. (2015) Resolving evolutionary relationships in closely related species with whole-genome sequencing data. Systematic Biology, 64 (6), 1017–2015. https://doi.org/10.1093/sysbio/syv045
- Palstra, A. P., de Graaf, M., & Sibbing, F. A. (2004) Riverine spawning and reproductive segregation in a lacustrine cyprinid species flock, facilitated by homing? Animal Biology, 54, 393–415. https://doi.org/10.1163/1570756042729519
- Paradis, E., Claude, J. & Strimmer, K. (2003) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics, 20, 289–290. https://doi.org/10.1093/bioinformatics/btg412
- Petren, K., Grant, P.R., Grant, B.R. & Keller, L.F. (2005) Comparative landscape genetics and the adaptive radiation of Darwin's finches: the role of peripheral isolation. Molecular Ecology, 14, 2943–2957. https://doi.org/10.1111/j.1365-294X.2005.02632.x
- Prave, A.R., Bates, C.R., Donaldson, C.H., Toland, H., Condon, D.J., Mark, D. & Raub, T.D. (2016) Geology and geochronology of the Tana Basin, Ethiopia: LIP volcanism, super eruptions and Eocene–Oligocene environmental change. Earth Planet. Science Letters, 443, 1–8. https://doi.org/10.1016/j.epsl.2016.03.009
- Pritchard, J. K., Stephens, M. & Donnely, P. (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. https://doi.org/10.1093/genetics/155.2.945
- Riesch, R., Muschick, M., Lindtke, D., Villoutreix, R., Comeault, A.A., Farkas, T.E., Lucek, K., Hellen, E., Soria-Carrasco, V., Dennis, S.R., de Carvalho, C.F., Safran, R.J., Sandoval, C.P., Feder, J., Gries, R., Crespi, B.J., Gries, G., Gompert, Z. & Nosil, P. (2017) Transitions between phases of genomic differentiation during stick-insect speciation. Nature Ecology & Evolution, 1, 0082. https://doi.org/0.1038/s41559-017-0082
- Rosenberg, N.A. (2004) DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
- Rüppell, W.P.E.S. (1836) Neuer Nachtrag von Beschreibungen und Abbildungen neuer Fische, im Nil entdeckt. Museum Senckenbergianum: Abhandlungen aus dem Gebiete der beschreibenden Naturgeschichte, van Mitgliedern der Senckenbergischen Naturforschenden Gesellschaft in Frankfurt, Main Frankfort, 2, 1–28.
- Salzburger, W. & Meyer, A. (2004) The species flock of east Africa cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften, 91, 277–290. https://doi.org/10.1007/s00114-004-0528-6
- Scrucca, L., Fop, M., Murphy, T.B. & Raftery, A.E. (2016) mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. The R Journal, 8 (1), 289–317. https://doi.org/10.32614/RJ-2016-021
- Seehausen, O. (2004) Hybridization and adaptive radiation. Trends in Ecology and Evolution, 19, 198–207. https://doi.org/10.1016/j.tree.2004.01.003
- Shaffer, H.B. & Thomson, R.C. (2007) Delimiting Species in Recent Radiations. Systematic Biology, 56, 896–906. https://doi.org/10.1080/10635150701772563
- Slatkin, M. (1994) Gene Flow and Population Structure. In: Real, L. Eds., Ecological Genetics, Princeton University Press, Princeton, 3–17.
- Soltis, P.S. & Soltis, D.E. (2000) The role of genetic and genomic attributes in the success of polyploids. PNAS, 97(13). 7051–7057. https://doi.org/10.1073/pnas.97.13.7051
- Süsnik, S., Snoj, A., Wilson, I.F., Mrdak, D. & Weiss, S. (2007). Historical demography of brown trout (Salmo trutta) in the Adriatic drainage including the putative S. letnica endemic to Lake Ohrid. Molecular Phylogenetics and Evolution, 44, 63–76. https://doi.org/10.1016/j.ympev.2006.08.021
- Vreven, E.J.W.M.N., Musschoot, T., Snoeks, J. & Schliewen, U.K. (2016) The African hexaploid Torini (Cypriniformes: Cyprinidae): review of a tumultuous history. Zoological Journal of Linnean Society, 177, 231–305. https://doi.org/10.1111/zoj.12366
- Wang, Y.C., Liao, L. & Li, Z.Z. (2018) Genetic differentiation of Actinidia chinensis and analysis of gene flow barriers in the Qinling Mountains, the species’ northern distribution boundary. Genetic Resources and Crop Evolution, 65, 881–895. https://doi.org/10.1007/s10722-017-0578-1
- Wang, Z., Hu, G., Li, Z., Zhong, C. & Yao, X. (2022) Characterizing Tetraploid populations of Actinidia chinensis for Kiwifruit genetic improvement. Plants, 11, 1154. https://doi.org/10.3390/plants11091154
- Wright, S. (1978) Evolutiom and the genetics of populations. Variability within and among natural populations (Vol. 4). University of Chicago Press, Chicago, IL, 590 pp.
- Zink, R.M. (1997) Phylogeographic studies of North American birds. In: Mindell, D.P. (Ed.), Avian Molecular Evolution and Systematics. Academic Press, San Diego, California, pp. 297–230. https://doi.org/10.1016/B978-012498315-1/50017-0
- Zink, R.M. & Barrowclough, G.F. (2008) Mitochondrial DNA under siege in avian phylogeography. Molecular Ecology, 17, 2107–2121. https://doi.org/10.1111/j.1365-294X.2008.03737.x
- Zworykin, D., Budaev, S., Darkov, A., Dzerzhinskii, K., Lyovin, B. & Mina, M. (2006) Assessment of the role of chemoreception in the mate choice in barbs of the Barbus intermedius; complex from Lake Tana, Ethiopia. Journal of Ichthyology, 46, 661–667. https://doi.org/10.1134/S0032945206080133