Abstract
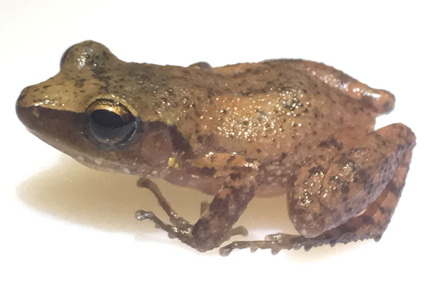
The number of species of the subgenus Syrrhophus, genus Eleutherodactylus has increased rapidly in the last eight years, due to recent taxonomic studies. This subgenus of direct-developing frogs is well represented in Mexico, which harbors more than 90% of the species richness reported for the subgenus. In this study, we describe one new species, Eleutherodactylus (Syrrhophus) coelum sp. nov. from the El Cielo Biosphere Reserve in Tamaulipas, Mexico, based on morphological and molecular data. The new species, which was previously assigned to E. cystignathoides, differs from its congeners mainly by its small size and by having the tips of Finger III and IV expanded to twice the width of the narrowest part of these fingers. Phylogenetic analyses recovered a well-supported relationship of the new species as the sister group to an undescribed taxon from San Luis Potosí. This analysis also indicates there are more lineages wating for description.
References
- Cruz-Elizalde, R., Ochoa-Ochoa, L., Flores-Villela, O. & Velasco, J.A. (2022) Taxonomic distinctiveness and phylogenetic variability of amphibians and reptiles in the cloud forest of Mexico. Community Ecology, 23, 87–102. https://doi.org/10.1007/s42974-022-00075-w
- Devitt, T.J., Tseng, K., Taylor-Adair, M., Koganti, S., Timugura, A. & Cannatella, D.C. (2023) Two new species of Eleutherodactylus from western and central Mexico (Eleutherodactylus jamesdixoni sp. nov., Eleutherodactylus humboldti sp. nov.). PeerJ, 11, e14985. https://doi.org/10.7717/peerj.14985
- Figueroa-Rangel, B.L., Willis, K.J. & Olvera-Vargas, M. (2012) Late-Holocene successional dynamics in a transitional forest of west-Central Mexico. The Holocene, 22, 143–153. https://doi.org/10.1177/0959683611414929
- Fouquet, A., Gilles, A., Vences, M., Marty, C., Blanc, M. & Gemmell, N.J. (2007) Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS One, 2, e1109. https://doi.org/10.1371/journal.pone.0001109
- Frost, D.R. (2024) Amphibian Species of the World: an Online Reference. Version 6.2. American Museum of Natural History, New York, New York. Available from: https://amphibiansoftheworld.amnh.org/index.php (accessed 29 March 2024)
- Grünwald, C.I., Reyes-Velasco, J., Franz-Chávez, H., Morales-Flores, K.I., Ahumada-Carrillo, I.T., Jones, J.M. & Boissinot, S. (2018) Six new species of Eleutherodactylus (Anura: systematic relationships and the validity of related species. Mesoamerican Herpetology, 5, 6–83.
- Grünwald, C., Reyes-Velasco, J., Franz-Chávez, H., Morales-Flores, K., Ahumada-Carrillo, I.T., Rodríguez, C.M. & Jones, J.M. (2021) Two new species of Eleutherodactylus (Anura: Eleutherodactylidae) from Southern Mexico, with comments on the taxonomy of related species and their advertisement calls. Amphibian & Reptile Conservation, 15, 1–35.
- Grünwald, C.I., Montaño-Ruvalcaba, C., Jones, J.M., Ahumada-Carrillo, I., Grünwald, A.J., Zheng, J., Strickland, J.L. & Reyes-Velasco, J. (2023) A novel species of piping frog Eleutherodactylus (Anura, Eleutherodactylidae) from southern Mexico. Herpetozoa, 36, 95–111. https://doi.org/10.3897/herpetozoa.36.e104707
- Han, X. & Fu, J. (2013) Does life history shape sexual size dimorphism in anurans? A comparative analysis. BMC Evolutionary Biology, 13, 1–11. https://doi.org/10.1186/1471-2148-13-27
- Hedges, S.B., Duellman, W.E. & Heinicke, M.P. (2008) New World direct-developing frogs (Anura: Terrarana): molecular phylogeny, classification, biogeography, and conservation. Zootaxa, 1737 (1), 1–182. https://doi.org/10.11646/zootaxa.1737.1.1
- Hernández-Austria, R., García-Vázquez, U.O., Grünwald, C.I. & Parra-Olea, G. (2022) Molecular phylogeny of the subgenus Syrrhophus (Amphibia: Anura: Eleutherodactylidae), with the description of a new species from Eastern Mexico. Systematics and Biodiversity, 20, 1–20. https://doi.org/10.1080/14772000.2021.2014597
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35, 518–522. https://doi.org/10.1093/molbev/msx281
- Huelsenbeck, J.P. & Rannala, B. (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology, 53, 904–913. https://doi.org/10.1080/10635150490522629
- Hutter, C.R., Andriampenomanana, Z.F., Andrianasolo, G.T., Cobb, K.A., Razafindraibe, J.H., Abraham, R.K. & Lambert, S.M. (2021) A fantastic new species of secretive forest frog discovered from forest fragments near Andasibe, Madagascar. Zoosystematics and Evolution, 97 (2), 483–495. https://doi.org/10.3897/zse.97.73630
- IUCN Standards and Petitions Committee. (2022) Guidelines for Using the IUCN Red List Categories and Criteria. Version 15.1. Prepared by the Standards and Petitions Committee. Available from: https://www.iucnredlist.org/documents/RedListGuidelines.pdf (accessed 25 September 2023)
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualisation. Briefings in Bioinformatics, 20, 1160–1166. https://doi.org/10.1093/bib/bbx108
- Köhler, G. (2012) Color Catalogue for Field Biologists. Bilingual Edition: English/Español. Herpeton Elke Kohler, Offenbach, 49 pp.
- Lanfear, R., Calcott, B., Ho, S.Y. & Guindon, S. (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29, 1695–1701. https://doi.org/10.1093/molbev/mss020
- Lanfear, R., Frandsen, P.B., Wright, A.M., Senfeld, T. & Calcott, B. (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34, 772–773. https://doi.org/10.1093/molbev/msw260
- Lynch, J.D. (1970) A taxonomic revision of the leptodactylid frog genus Syrrhophus Cope. University of Kansas Publications Museum of Natural History 20. University of Kansas, Lawrence, Kansas, 48 pp. https://doi.org/10.5962/bhl.part.2809
- Lynch, J.D. & Duellman, W.E. (1980) The Eleutherodactylus of the Amazonian Slopes of the Ecuadorian Andes (Anura: Leptodactylidae). The University of Kansas, Museum of Natural History, Miscellaneous Publication 69. The University of Kansas, Lawrence, Kansas, 86 pp. https://doi.org/10.5962/bhl.title.16222
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 gateway computing environments workshop (GCE), 2010, 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Minh, B.Q., Nguyen, M.A.T. & Von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30, 1188–1195. https://doi.org/10.1093/molbev/mst024
- Müller, K., Müller, J., Neinhuis, C. & Quandt, D. (2010) PhyDE—Phylogenetic Data Editor. Version 0.9971. Program distributed by the authors. Available from: http:www.Phyde.de (accessed 29 May 2024)
- Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
- Oksanen, J., Simpson, G., Blanchet, F., Kindt, R., Legendre, P., Minch’n, P., O'Hara, R., Solymos, P., Stevens, M., Szoecs, E., Wagner, H., Barbour, M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M., De Caceres, M., Durand, S., Evangelista, H., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M., Lahti, L., McGlinn, D., Ouellette, M., Ribeiro Cunha, E., Smith, T., Stier, A., Ter Braak, C. & Weedon, J. (2022) vegan: Community Ecology Package. R package. Version 2.6-4. Available from: https://CRAN.R-project.org/package=vegan (accessed 3 April 2024)
- R Core Team (2023) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: https://www.R-project.org/ (accessed 3 April 2024)
- Palacios-Aguilar, R. & Santos-Bibiano, R. (2020) A new species of direct-developing frog of the genus Eleutherodactylus (Anura: Eleutherodactylidae) from the Pacific lowlands of Guerrero, Mexico. Zootaxa, 4750 (2), 250–260. https://doi.org/10.11646/zootaxa.4750.2.8
- Palumbi, S.R., Martin, A., Romano, S., McMillan, W.O., Stice, L. & Grabowski, G. (1991) The simple fool’s guide to PCR. Version 2.0. University of Hawaii, Honolulu, 45 pp.
- Rambaut, A., Drummond, A.J., Xie, D., Baele, G. & Suchard, M.A. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67, 901–904. https://doi.org/10.1093/sysbio/syy032
- Reyes-Velasco, J., Ahumada-Carrillo, I., Burkhardt, T.R. & Devitt, T.J. (2015) Two new species of Eleutherodactylus (subgenus Syrrhophus) from western Mexico. Zootaxa, 3914 (3), 301–317. https://doi.org/10.11646/zootaxa.3914.3.4
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
- Rovito, S.M. & Parra-Olea, G. (2015) Two new species of Chiropterotriton (Caudata: Plethodontidae) from northern Mexico. Zootaxa, 4048 (1), 57–74. https://doi.org/10.11646/zootaxa.4048.1.3
- Shimada, T., Matsui, M., Yambun, P., Lakim, M. & Mohamed, M. (2008) Detection of two cryptic taxa in Meristogenys amoropalamus (Amphibia, Ranidae) through nuclear and mitochondrial DNA analyses. Zootaxa, 1843 (1), 24–34. https://doi.org/10.11646/zootaxa.1843.1.2
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution, 38, 3022–3027. https://doi.org/10.1093/molbev/msab120
- Trifinopoulos, J., Nguyen, L.T., Von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44, W232–W235. https://doi.org/10.1093/nar/gkw256
- Sambrook, J. & Russell, D.W. (2006) Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harbor Protocols, 2006. [published online] https://doi.org/10.1101/pdb.prot4455
- Shine, R. (1979) Sexual selection and sexual dimorphism in the Amphibia. Copeia, 1979, 297–306. https://doi.org/10.2307/1443418
- Soto-Pozos, A.F., Basanta, M.D., García-Castillo, M.G. & Parra-Olea, G. (2023) The Amphibians of the Mexican Montane Cloud Forest. In: Jones, R.W., Ornelas-García, C.P., Pineda-López, R. & Álvarez, F. (Eds.), Mexican Fauna in the Anthropocene. Springer International Publishing, Cham, pp. 357–376. https://doi.org/10.1007/978-3-031-17277-9_17
- Vallejo-Pareja, M.C., Stanley, E.L., Bloch, J.I. & Blackburn, D.C. (2023) Fossil frogs (Eleutherodactylidae: Eleutherodactylus) from Florida suggest overwater dispersal from the Caribbean by the Late Oligocene. Zoological Journal of the Linnean Society, 201 (2), 431–446. https://doi.org/10.1093/zoolinnean/zlad130