Abstract
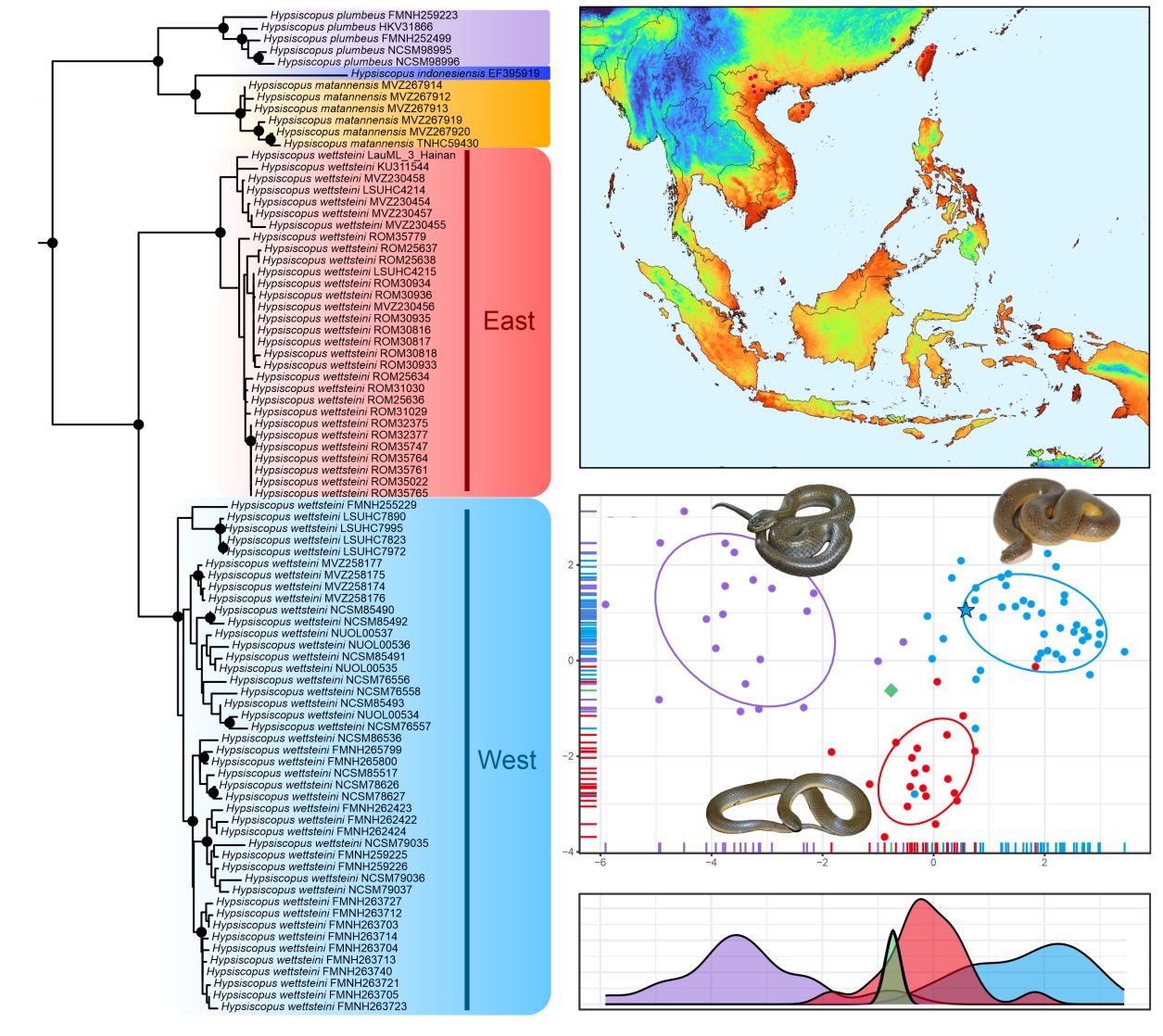
Homalopsids (Old World Mud Snakes) include 59 semiaquatic species in Asia and Australasia that display an array of morphological adaptations, behaviors, and microhabitat preferences. These attributes make homalopsids an ideal model system for broader questions in evolutionary biology, but the diversity of this understudied group of snakes is still being described. Recognized species diversity in rice paddy snakes (Hypsiscopus) has recently doubled after nearly 200 years of taxonomic stability. However, the evolutionary distinctiveness of some populations remains in question. In this study, we compare mainland Southeast Asian populations of Hypsiscopus east and west of the Red River Basin in Vietnam, a known biogeographic barrier in Asia, using an iterative approach with molecular phylogenetic reconstruction, machine-learning morphological quantitative statistics, and ecological niche modeling. Our analyses show that populations west of the Red River Basin represent an independent evolutionary lineage that is distinct in genetics, morphospace, and habitat suitability, and so warrants species recognition. The holotype of H. wettsteini, a species originally described in error from Costa Rica, grouped morphometrically with the population at the Red River Basin and eastward, and those west of the Red River Basin are referred to the recently described H. murphyi. The two species may have diversified due to a variety of geological and environmental factors, and their recognition exemplifies the importance of multifaceted approaches in taxonomy for downstream biogeographic studies on speciation scenarios.
References
- Ahmadzadeh, F., Flecks, M., Carretero, M.A., Böhme, W., Ilgaz, C., Engler, J.O., James Harris, D., Üzüm, N. & Rödder, D. (2013) Rapid lizard radiation lacking niche conservatism: ecological diversification within a complex landscape. Journal of Biogeography, 40, 1807–1818. https://doi.org/10.1111/jbi.12121
- Aiello-Lammens, M.E., Boria, R.A., Radosavljevic, A., Vilela, B. & Anderson, R.P. (2015) spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38, 541–545. https://doi.org/10.1111/ecog.01132
- Bain, R.H. & Hurley, M.M. (2011) A biogeographic synthesis of the amphibians and reptiles of Indochina. Bulletin of the American Museum of Natural History, 2011, 1–138. https://doi.org/10.1206/360.1
- Becker, R.A., Wilks, A.R., Brownrigg, R., Minka, T.P. & Deckmyn, A. (2018) Maps: Draw Geographical Maps. Available from: https://cran.r-project.org/web/packages/maps/maps.pdf (accessed 29 February 2024)
- Bernstein, J.M., Murphy, J.C., Voris, H.K., Brown, R.M. & Ruane, S. (2021) Phylogenetics of mud snakes (Squamata: Serpentes: Homalopsidae): A paradox of both undescribed diversity and taxonomic inflation. Molecular Phylogenetics and Evolution, 160, 107109. https://doi.org/10.1016/j.ympev.2021.107109
- Bernstein, J.M. & Ruane, S. (2022) Maximizing molecular data from low-quality fluid-preserved specimens in natural history collections. Frontiers in Ecology and Evolution, 10, 893088. https://doi.org/10.3389/fevo.2022.893088
- Bernstein, J.M., de Souza, H.F., Murphy, J.C., Voris, H.K., Brown, R.M., Myers, E.A., Harrington, S., Shanker, K. & Ruane, S. (2023) Phylogenomics using fresh and formalin specimens resolves the systematics of Old World Mud Snakes (Serpentes: Homalopsidae) and expands biogeographic inference. Bulletin of the Society of Systematic Biologists, 2 (1), 1–24. https://doi.org/10.18061/bssb.v2i1.9393
- Bernstein, J.M., Voris, H.K., Stuart, B.L., Karns, D.R., McGuire, J.A., Iskandar, D.T., Riyanto, A., Calderón-Acevedo, C.A., Brown, R.M., Gehara, M., Soto-Centeno, J.A. & Ruane, S. (2024) Integrative methods reveal multiple drivers of diversification in rice paddy snakes. Scientific Reports, 14, 4727. https://doi.org/10.1038/s41598-024-54744-z
- Bernstein, J.M., Voris, H.K., Stuart, B.L., Phimmachak, S., Seateun, S., Sivongxay, N., Neang, T., Karns, D.R., Andrews, H.L., Osterhage, J., Phipps, E.A. & Ruane, S. (2022) Undescribed diversity in a widespread, common group of Asian mud snakes (Serpentes: Homalopsidae: Hypsiscopus). Ichthyology & Herpetology, 110 (3), 561–574. https://doi.org/10.1643/h2022015
- Bivand, R., Keitt, T., Rowlingson, B., Pebesma, E., Sumner, M., Hijmans, R., Baston, D., Rouault, E., Warmerdam, F., Ooms, J. & Rundel, C. (2021) rgdal: Bindings for the “Geospatial” Data Abstraction Library. Available from: https://rgdal.r-forge.r-project.org/ (accessed 29 February 2024)
- Bivand, R. & Lewin-Koh, N. (2021) maptools: Tools for Handling Spatial Objects. Available from: https://maptools.r-forge.r-project.org/ (accessed 29 February 2024)
- Breitfeld, H.T., Hennig-Breitfeld, J., BouDagher-Fadel, M.K., Hall, R. & Galin, T. (2020) Oligocene-Miocene drainage evolution of NW Borneo: Stratigraphy, sedimentology and provenance of Tatau-Nyalau province sediments. Journal of Asian Earth Sciences, 195, 104331. https://doi.org/10.1016/j.jseaes.2020.104331
- Burbrink, F.T., Bernstein, J.M., Kuhn, A., Gehara, M. & Ruane, S. (2021) Ecological divergence and the history of gene flow in the Nearctic milksnakes (Lampropeltis triangulum complex). Systematic Biology, 71 (4), 839–858. https://doi.org/10.1093/sysbio/syab093
- Catania, K.C., Leitch, D.B. & Gauthier, D. (2010) Function of the appendages in tentacled snakes ( Erpeton tentaculatus ). Journal of Experimental Biology, 213, 359–367. https://doi.org/10.1242/jeb.039685
- Chan, K.O., Sind, L.I., Thong, L.I., Ananthanarayanan, S., Rasu, S., Aowphol, A., Rujirawan, A., Anuar, S., Mulcahy, D., Grismer, J.L. & Grismer, L.L. (2022) Phylogeography of mangrove pit vipers (Viperidae, Trimeresurus erythrurus‐purpureomaculatus complex). Zoologica Scripta, 51, 664–675. https://doi.org/10.1111/zsc.12562
- Chen, D. & Chen, H.W. (2013) Using the Köppen classification to quantify climate variation and change: An example for 1901–2010. Environmental Development, 6, 69–79. https://doi.org/10.1016/j.envdev.2013.03.007
- Cox, M.J. (1991) Snakes of Thailand and their husbandry. Krieger Pub Co, Malabar, Florida, 526 pp.
- David, P. & Vogel, G. (2024) On the status of Helicops wettsteini Amaral, 1929, a senior synonym of Hypsiscopus murphyi (SERPENTES: Homalopsidae). Zootaxa, 5415 (2), 300–308. https://doi.org/10.11646/zootaxa.5415.2.4
- De Bruyn, M., Rüber, L., Nylinder, S., Stelbrink, B., Lovejoy, N.R., Lavoué, S., Tan, H.H., Nugroho, E., Wowor, D., Ng, P.K.L., Azizah, M.N.S., von Rintelen, T., Hall, R . & Carvalho, G. R. (2013) Paleo-drainage basin connectivity predicts evolutionary relationships across three Southeast Asian biodiversity hotspots. Systematic Biology, 62 (3), 398–410.
- De Queiroz, K. (2007) Species concepts and species delimitation. Systematic Biology, 56, 879–886. https://doi.org/10.1080/10635150701701083
- Di Cola, V., Broennimann, O., Petitpierre, B., Breiner, F.T., D’Amen, M., Randin, C., Engler, R., Pottier, J., Pio, D., Dubuis, A., Pellissier, L., Mateo, R.G., Hordijk, W., Salamin, N. & Guisan, A. (2017) ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography, 40, 774–787. https://doi.org/10.1111/ecog.02671
- Enriquez‐Urzelai, U., Martínez‐Freiría, F., Freitas, I., Perera, A., Martínez‐Solano, Í., Salvi, D., Velo‐Antón, G. & Kaliontzopoulou, A. (2022) Allopatric speciation, niche conservatism and gradual phenotypic change in the evolution of European green lizards. Journal of Biogeography, 49, 2193–2205. https://doi.org/10.1111/jbi.14497
- Fabre, A.-C., Bickford, D., Segall, M. & Herrel, A. (2016) The impact of diet, habitat use, and behaviour on head shape evolution in homalopsid snakes. Biological Journal of the Linnean Society, 118, 634–647. https://doi.org/10.1111/bij.12753
- Favre, A., Päckert, M., Pauls, S.U., Jähnig, S.C., Uhl, D., Michalak, I. & Muellner‐Riehl, A.N. (2015) The role of the uplift of the Qinghai‐Tibetan Plateau for the evolution of Tibetan biotas. Biological Reviews, 90, 236–253. https://doi.org/10.1111/brv.12107
- Fick, S.E. & Hijmans, R.J. (2017) WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37, 4302–4315. https://doi.org/10.1002/joc.5086
- Fu, J. & Wen, L. (2023) Impacts of Quaternary glaciation, geological history and geography on animal species history in continental East Asia: A phylogeographic review. Molecular Ecology, 32, 4497–4514. https://doi.org/10.1111/mec.17053
- Garnier, S., Ross, N., Rudis, R., Camargo, A.P., Sciaini, M. & Scherer, C. (2021) Rvision - Colorblind-Friendly Color Maps for R. R package version 0.6.2. Available from: https://sjmgarnier.github.io/viridis/authors.html (accessed 29 February 2024)
- Gressitt, J.L. (1941) Amphibians and reptiles from southeastern China. Philippine Journal of Science, 75, 1–58.
- Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. https://doi.org/10.1093/sysbio/syq010
- Hall, R. (1996) Reconstructing Cenozoic SE Asia. In: Hall, R. & Blundell, D. (Eds.), Tectonic Evolution of Southeast Asia. Geological Society of London, London, pp. 153–184.
- Hall, R. (1998) The plate tectonics of Cenzoic SE Asia and the distribution of land and sea. In: Hall, R. & Holloway, J.D. (Eds.), Biogeography and geological Evolution of SE Asia. Backbuys Publishers, Leiden, pp. 99–131.
- Hall, R. (2009) Southeast Asia’s changing palaeogeography. Blumea - Biodiversity, Evolution and Biogeography of Plants, 54, 148–161. https://doi.org/10.3767/000651909X475941
- Hamidy, A., Zakky, Q., Fitriyana, N. & Endarwin, W. (2023) A new species of water snake genus Hypsiscopus (Serpentes: Homalopsidae) from Sulawesi, Indonesia. TREUBIA, 50, 21–38. https://doi.org/10.14203/treubia.v50i1.4511
- Hijmans, R.J., Etten, J. van, Sumner, M., Cheng, J., Baston, D., Bevan, A., Bivand, R., Busetto, L., Canty, M., Fasoli, B., Forrest, D., Ghosh, A., Golicher, D., Gray, J., Greenberg, J.A., Hiemstra, P., Hingee, K., Ilich, A., Geosciences, I. for M.A., Karney, C., Mattiuzzi, M., Mosher, S., Naimi, B., Nowosad, J., Pebesma, E., Lamigueiro, O.P., Racine, E.B., Rowlingson, B., Shortridge, A., Venables, B. & Wueest, R. (2022) raster: Geographic Data Analysis and Modeling. Available from: https://cran.r-project.org/web/packages/raster/index.html (accessed 29 February 2024)
- Hijmans, R.J. & Graham, C.H. (2006) The ability of climate envelope models to predict the effect of climate change on species distributions: comparing climate envelope and mechanistic models. Global Change Biology, 12, 2272–2281. https://doi.org/10.1111/j.1365-2486.2006.01256.x
- Hijmans, R.J., Phillips, S. & Elith, J.L.J. (2021) dismo: Species Distribution Modeling. Available from: https://cran.r-project.org/web/packages/dismo/index.html (accessed 29 February 2024)
- Hua, X. & Wiens, J.J. (2010) Latitudinal variation in speciation mechanisms in frogs. Evolution, 64, 429–443. https://doi.org/10.1111/j.1558-5646.2009.00836.x
- Hutchison, C.S. (1989) Geological Evolution of South-east Asia. 2nd Edition. Claredon Press, Oxford, 406 pp.
- Jayne, B.C., Voris, H.K. & Ng, P.K.L. (2018) How big is too big? Using crustacean-eating snakes (Homalopsidae) to test how anatomy and behaviour affect prey size and feeding performance. Biological Journal of the Linnean Society, 123, 636–650. https://doi.org/10.1093/biolinnean/bly007
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
- Karin, B.R., Gamble, T. & Jackman, T.R. (2020) Optimizing phylogenomics with rapidly evolving long exons: comparison with anchored hybrid enrichment and ultraconserved elements. Molecular Biology and Evolution, 37, 904–922. https://doi.org/10.1093/molbev/msz263
- Karns, D.R., Lukoschek, V., Osterhage, J., Murphy, J.C. & Voris, H.K. (2010) Phylogeny and biogeography of the Enhydris clade (Serpentes: Homalopsidae). Zootaxa, 2452 (1), 18–30. https://doi.org/10.11646/zootaxa.2452.1.2
- Karsen, S.J., Wai-neng Lau, M. & Bogadek, A. (1986) Hong Kong Amphibians and Reptiles. Provisional Urban Council Publication, Hong Kong, 136 pp.
- Kass, J.M., Muscarella, R., Galante, P.J., Bohl, C.L., Pinilla-Buitrago, G.E., Boria, R.A., Soley-Guardia, M. & Anderson, R.P. (2021) ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods in Ecology and Evolution, 12, 1602–1608. https://doi.org/10.1111/2041-210X.13628
- Knouft, J.H., Losos, J.B., Glor, R.E. & Kolbe, J.J. (2006) Phylogenetic analysis of the evolution of the niche in lizards of the Anolis sagrei group. Ecology, 87, S29–S38. https://doi.org/10.1890/0012-9658(2006)87[29:PAOTEO]2.0.CO;2
- Kunts, R.E. (1983) Snakes of Taiwan. United States Navy Medical Research Unit No. 2, Taipei, 44 pp.
- Lamigueiro, O.P. & Hijmans, R. (2022) rasterVis: Visualization Methods for Raster Data. Available from: https://cran.r-project.org/web/packages/rasterVis/index.html (accessed 29 February 2024)
- Lanfear, R., Frandsen, P.B., Wright, A.M., Senfeld, T . & Calcott, B. (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular biology and evolution, 34 (3), 772–773.
- Leloup, P.H., Lacassin, R., Tapponnier, P., Schärer, U., Zhong, D., Liu, X., Zhang, L., Ji, S. & Trinh, P.T. (1995) The Ailao Shan-Red River shear zone (Yunnan, China), Tertiary transform boundary of Indochina. Tectonophysics, 251, 3–84. https://doi.org/10.1016/0040-1951(95)00070-4
- Li, J.-N., He, C., Guo, P., Zhang, P. & Liang, D. (2017) A workflow of massive identification and application of intron markers using snakes as a model. Ecology and Evolution, 7, 10042–10055. https://doi.org/10.1002/ece3.3525
- Li, J.-N., Liang, D., Wang, Y.-Y., Guo, P., Huang, S. & Zhang, P. (2020) A large-scale systematic framework of Chinese snakes based on a unified multilocus marker system. Molecular Phylogenetics and Evolution, 148, 106807. https://doi.org/10.1016/j.ympev.2020.106807
- Mell, R. (1922) Beiträge zur Fauna sinica. Die Vertebraten Südchinas: Feldlisten und Feldnoten der Säuger, Vögel, Reptilien, Batrachier, 122 pp.
- Minh, B.Q., Nguyen, M.A.T. & von Haeseler, A. (2013) Ultrafast Approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30, 1188–1195. https://doi.org/10.1093/molbev/mst024
- Mulch, A. & Chamberlain, C.P. (2006) The rise and growth of Tibet. Nature, 439, 670–671. https://doi.org/10.1038/439670a
- Muñoz, M.M., Crawford, N.G., McGreevy, T.J., Messana, N.J., Tarvin, R.D., Revell, L.J., Zandvliet, R.M., Hopwood, J.M., Mock, E., Schneider, A.L. & Schneider, C.J. (2013) Divergence in coloration and ecological speciation in the Anolis marmoratus species complex. Molecular Ecology, 22, 2668–2682. https://doi.org/10.1111/mec.12295
- Murphy, J.C. (2007) Homalopsid Snakes: Evolution in the Mud. Krieger Publishing Company, Malabar, 260 pp.
- Murphy, J.C. & Voris, H.K. (2014) A checklist and key to the homalopsid snakes (Reptilia, Squamata, Serpentes), with the description of new genera. Fieldiana Life and Earth Sciences, 2014, 1–43. https://doi.org/10.3158/2158-5520-14.8.1
- Murphy, J.C. & Voris, H.K. (2021) A new species of Brachyorrhos from Seram, Indonesia and notes on fangless homalopsids (Squamata, Serpentes). Philippine Journal of Systematic Biology, 14 (2), 1–8 + i–ii. https://doi.org/10.26757/pjsb2020b14015
- Murphy, J.C., Voris, H.K. & Karns, D.R. (2012a) The dog-faced water snakes, a revision of the genus Cerberus Cuvier, (Squamata, Serpentes, Homalopsidae), with the description of a new species. Zootaxa, 3484 (1), 1–34. https://doi.org/10.11646/zootaxa.3484.1.1
- Murphy, J.C., Voris, H.K., Karns, D.R., Chan-ard, T. & Suvunrat, K. (1999) The ecology of the water snakes of Ban Tha Hin, Songkhla Province, Thailand. Natural History Bulletin of the Siam Society, 47, 129–147.
- Murphy, J.C., Voris, H.K., Murthy, B.H.C.K., Traub, J. & Cumberbatch, C. (2012b) The masked water snakes of the genus Homalopsis Kuhl & van Hasselt, 1822 (Squamata, Serpentes, Homalopsidae), with the description of a new species. Zootaxa, 3208 (1), 1–26. https://doi.org/10.11646/zootaxa.3208.1.1
- Neuwirth, E. (2022) RColorBrewer: ColorBrewer Palettes. Available from: https://r-graph-gallery.com/38-rcolorbrewers-palettes.html (accessed 29 February 2024)
- Nguyen, L.-T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
- O’Connell, K.A., Mulder, K.P., Wynn, A., Queiroz, K. & Bell, R.C. (2021) Genomic library preparation and hybridization capture of formalin‐fixed tissues and allozyme supernatant for population genomics and considerations for combining capture‐ and RADseq‐based single nucleotide polymorphism data sets. Molecular Ecology Resources, 22 (2), 487–502. https://doi.org/10.1111/1755-0998.13481
- Ogden, R. & Thorpe, R.S. (2002) Molecular evidence for ecological speciation in tropical habitats. Proceedings of the National Academy of Sciences, 99, 13612–13615. https://doi.org/10.1073/pnas.212248499
- Pebesma, E. (2018) Simple Features for R: Standardized Support for Spatial Vector Data. The R Journal, 10, 439. https://doi.org/10.32614/RJ-2018-009
- Peel, M.C., Finlayson, B.L. & McMahon, T.A. (2007) Updated world map of the Köppen-Geiger climate classification. Hydrology and Earth System Sciences, 11, 1633–1644. https://doi.org/10.5194/hess-11-1633-2007
- Phillips, S.J., Anderson, R.P. & Schapire, R.E. (2006) Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190, 231–259. https://doi.org/10.1016/j.ecolmodel.2005.03.026
- Phillips, S.J., Dudík, M. & Schapire, R.E. (2004) A maximum entropy approach to species distribution modeling. In: Proceedings of the twenty-first international conference on Machine learning. ICML ’04. Association for Computing Machinery, New York, New York, pp. 83. https://doi.org/10.1145/1015330.1015412
- Pope, C.H. (1929) A list of reptiles known to occur in Fukien Province, China. Proceedings of the Natural History Society of Fukien, 2, 20–22.
- Pyron, A.R. & Burbrink, F.T. (2009) Lineage diversification in a widespread species: roles for niche divergence and conservatism in the common kingsnake, Lampropeltis getula. Molecular Ecology, 18, 3443–3457. https://doi.org/10.1111/j.1365-294X.2009.04292.x
- Qu, Y., Song, G., Gao, B., Quan, Q., Ericson, P.G.P. & Lei, F. (2015) The influence of geological events on the endemism of East Asian birds studied through comparative phylogeography B. Riddle (Ed). Journal of Biogeography, 42, 179–192. https://doi.org/10.1111/jbi.12407
- Quah, E.S.H., Grismer, L.L., Wood, P.L., Jr., Thura, M.K., Zin, T., Kyaw, H., Lwin, N., Grismer, M.S. & Murdoch, M.L. (2017) A new species of Mud Snake (Serpentes, Homalopsidae, Gyiophis Murphy & Voris, 2014) from Myanmar with a first molecular phylogenetic assessment of the genus. Zootaxa, 4238 (4), 571–582. https://doi.org/10.11646/zootaxa.4238.4.5
- Quah, E.S.H., Wood, P.L.Jr., Grismer, L.L. & Sah, S.A.M. (2018) On the taxonomy and phylogeny of the rare Selangor Mud Snake (Raclitia indica) Gray (Serpentes, Homalopsidae) from Peninsular Malaysia. Zootaxa, 4514 (1), 53. https://doi.org/10.11646/zootaxa.4514.1.4
- Rambaut, A. (2014) FigTree v1.4.2. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 29 February 2024)
- Ramos, E.K.S., Magalhães, R.F. de, Marques, N.C.S., Baêta, D., Garcia, P.C.A. & Santos, F.R. (2019) Cryptic diversity in Brazilian endemic monkey frogs (Hylidae, Phyllomedusinae, Pithecopus) revealed by multispecies coalescent and integrative approaches. Molecular Phylogenetics and Evolution, 132, 105–116. https://doi.org/10.1016/j.ympev.2018.11.022
- Raxworthy, C.J., Ingram, C.M., Rabibisoa, N. & Pearson, R.G. (2007) Applications of ecological niche modeling for species delimitation: a review and empirical evaluation using day geckos (Phelsuma) from Madagascar. Systematic Biology, 56, 907–923. https://doi.org/10.1080/10635150701775111
- Richards, C.L., Carstens, B.C. & Lacey Knowles, L. (2007) Distribution modelling and statistical phylogeography: an integrative framework for generating and testing alternative biogeographical hypotheses. Journal of Biogeography, 34, 1833–1845. https://doi.org/10.1111/j.1365-2699.2007.01814.x
- Rissler, L.J. & Apodaca, J.J. (2007) Adding more ecology into species delimitation: ecological niche models and phylogeography help define cryptic species in the Black Salamander (Aneides flavipunctatus). Systematic Biology, 56, 924–942. https://doi.org/10.1080/10635150701703063
- Ronquist, F.M., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A . & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61 (3), 539–542. https://doi.org/10.1093/sysbio/sys029
- Rossman, D.A. & Scott, N. J. (1968) Identity of Helicopes wettsteini Amaral (Serpentes: Colubridae). Herpetologica, 24 (3), 262–263.
- Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J.C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S.E. & Sánchez-Gracia, A. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299–3302. https://doi.org/10.1093/molbev/msx248
- Ruane, S. & Austin, C.C. (2017) Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Molecular Ecology Resources, 17, 1003–1008. https://doi.org/10.1111/1755-0998.12655
- Sabaj, M.H. (2016) Standard symbolic codes for institutional resource collections in herpetology and ichthyology: an online reference. Version 6.5. American Society of Ichthyologists and Herpetologists, 108, 593–669. https://doi.org/10.1643/ASIHCODONS2020
- Salles, T., Mallard, C., Husson, L., Zahirovic, S., Sarr, A.-C. & Sepulchre, P. (2021) Quaternary landscape dynamics boosted species dispersal across Southeast Asia. Communications Earth & Environment, 2, 240. https://doi.org/10.1038/s43247-021-00311-7
- Schmidt, K.P. (1927) The reptiles of Hainan. Bulletin of the American Museum of Natural History, 54, 395–465.
- Schneider, C.J., Smith, T.B., Larison, B. & Moritz, C. (1999) A test of alternative models of diversification in tropical rainforests: Ecological gradients vs. rainforest refugia. Proceedings of the National Academy of Sciences, 96, 13869–13873. https://doi.org/10.1073/pnas.96.24.13869
- Shen, X.X., Liang, D., Feng, Y.J., Chen, M.Y. & Zhang, P. (2013) A versatile and highly efficient toolkit including 102 nuclear markers for vertebrate phylogenomics, tested by resolving the higher level relationships of the Caudata. Molecular Biology and Evolution, 30, 2235–2248. https://doi.org/10.1093/molbev/mst122
- Simmons, J.E. (2015) Herpetological collecting and collections management. Society for the Study of Amphibians and Reptiles Herpetological Circular, 42, 1–191.
- Soto-Centeno, J.A. (2022) ENMpipe: a tutorial pipeline for building and testing ecological niche models. Available from: https://github.com/mormoops/ENMpipe (accessed 29 February 2024)
- Urbanek, S. (2021) rJava: Low-Level R to Java Interface. Available from: https://cran.r-project.org/web/packages/rJava/index.html (accessed 29 February 2024)
- Voris, H.K. (2000) Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. Journal of Biogeography, 27, 1153–1167. https://doi.org/10.1046/j.1365-2699.2000.00489.x
- Weijola, V., Vahtera, V., Lindqvist, C. & Kraus, F. (2019) A molecular phylogeny for the Pacific monitor lizards (Varanus subgenus Euprepiosaurus) reveals a recent and rapid radiation with high levels of cryptic diversity. Zoological Journal of the Linnean Society, 186, 1053–1066. https://doi.org/10.1093/zoolinnean/zlz002
- Wickham, H. (2011) ggplot2: ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics, 3, 180–185. https://doi.org/10.1002/wics.147
- Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L.D., François, R., Grolemund, G., Hayes, A., Henry, L., Hester, J., Kuhn, M., Pedersen, T.L., Miller, E., Bache, S.M., Müller, K., Ooms, J., Robinson, D., Seidel, D.P., Spinu, V., Takahashi, K., Vaughan, D., Wilke, C., Woo, K. & Yutani, H. (2019) Welcome to the Tidyverse. Journal of Open Source Software, 4, 1686. https://doi.org/10.21105/joss.01686
- Wickham, H., François, R., Henry, K. & Müller, K. (2020) dplyr: a grammar of data manipulation. Available from: https://dplyr.tidyverse.org/ (accessed 29 February 2024)
- Wiens, J.J. (2004) Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution, 58, 193–197. https://doi.org/10.1111/j.0014-3820.2004.tb01586.x
- Wiens, J.J. & Graham, C.H. (2005) Niche conservatism: integrating evolution, ecology, and conservation biology. Annual Review of Ecology, Evolution, and Systematics, 36, 519–539. https://doi.org/10.1146/annurev.ecolsys.36.102803.095431
- Yuan, Z.-Y., Suwannapoom, C., Yan, F., Poyarkov, N.A., Nguyen, S.N., Chen, H., Chomdej, S., Murphy, R.W. & Che, J. (2016) Red River barrier and Pleistocene climatic fluctuations shaped the genetic structure of Microhyla fissipes complex (Anura: Microhylidae) in southern China and Indochina. Current Zoology, 62, 531–543. https://doi.org/10.1093/cz/zow042
- Zhang, D.-R., Chen, M.-Y., Murphy, R.W., Che, J., Pang, J.-F., Hu, J.-S., Luo, J., Wu, S.-J., Ye, H. & Zhang, Y.-P. (2010a) Genealogy and palaeodrainage basins in Yunnan Province: Phylogeography of the Yunnan spiny frog, Nanorana yunnanensis (Dicroglossidae). Molecular Ecology, 19, 3406–3420. https://doi.org/10.1111/j.1365-294X.2010.04747.x
- Zhang, M., Rao, D., Yang, J., Yu, G. & Wilkinson, J.A. (2010b) Molecular phylogeography and population structure of a mid-elevation montane frog Leptobrachium ailaonicum in a fragmented habitat of southwest China. Molecular Phylogenetics and Evolution, 54, 47–58. https://doi.org/10.1016/j.ympev.2009.10.019