Abstract
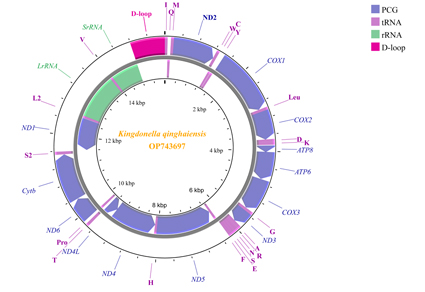
Kingdonella qinghaiensis Zheng, 1990 a species belonging to the genus Kingdonella Uvarov, 1933, within the family Acrididae in the order Orthoptera, is mainly distributed in Qinghai province, China. In this study, we determined, assembled and annotated the mitochondrial genome of Kingdonella qinghaiensis. The mitogenome is 15,597 bp in length and contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes and a control region (D-loop). The entire mitogenome exhibits a strong A/T bias, with an A+T content of 75.4%. All 13 PCGs had the typical start codon of ATN (9 ATGs, 2 ATCs and 2 ATTs) and TAA was the most frequent stop codon in Kingdonella qinghaiensis PCGs, except TAGs for ND3 and ND1 genes. A total of 3,730 codons are present in the mitogenomic PCGs of Kingdonella qinghaiensis. Among these, UUA (9.65%) is the most frequent codon for leucine (L), followed by AUU (9.3%) for isoleucine (I), and UUU (8.12%) for phenylalanine (F). The Ka/Ks ratios of the 13 PCGs in Kingdonella qinghaiensis mitogenome ranged from 0.1436 to 0.9107 (0<Ka/Ks<1), and ND4L had the highest ratio while COX1 gene had the lowest value. The nucleotides diversity (Pi) analysis of the 13 PCGs from 46 species in Acrididae implied that ND2 gene contained the highest variability site (0.27862). While ND5, ND4 and ND1 had comparatively low nucleotide diversities. The phylogenetic tree based on the nucleotide sequences of the 13 PCGs from 46 different species (including 2 outgroups) supported the monophyly of Acrididae and indicated two stable clades in Acrididae. The phylogenetic analyses represented the main topology as follows: ((((Catantopinae+Calliptaminae) +Cyrtacanthacridinae) + ((Spathosterninae+Oxyinae) + Melanoplinae)) + (((Oedipodinae+Acridinae) + Gomphocerinae) +Coptacrinae)). Species from Melanoplinae in the phylogenetic tree confirmed that Kingdonella qinghaiensis had the closer taxonomy relationship with Kingdonella bicollina, another member in the same genus.
References
- Avise, J.C. (1993) Molecular markers, natural history and evolution. Systematic Biology, 44 (1), 511. https://doi.org/10.1007/978-1-4615-2381-9_3
- Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., Pütz, J., Middendorf, M. & Stadler, P.F. (2013) MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Molecular Phylogenetics and Evolution, 69 (2), 313–319. https://doi.org/10.1016/j.ympev.2012.08.023
- Cameron, S.L. (2014) Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annual Review of Entomology, 59, 95–117. https://doi.org/10.1146/annurev-ento-011613-162007
- Chen, X., Yuan, Z., Li, C., Dietrich, C.H. & Song, Y. (2021) Structural features and phylogenetic implications of Cicadellidae subfamily and two new mitogenomes leafhoppers. PLoS ONE, 16 (5), e0251207. https://doi.org/10.1371/journal.pone.0251207
- Cigliano, M.M., Braun, H., Eades, D.C. & Otte, D. (2022) Orthoptera species file. Version 5.0/5.0. Available from: http://Orthoptera.SpeciesFile.org (accessed 21 October 2024)
- Curle, J.P. & Kocher, T.D. (1999) Mitogenomics: digging deeper with complete mitochondrial genomes. Trends in Ecology & Evolution, 47, 311–335. https://doi.org/10.1016/S0169-5347(99)01660-2
- da Silva, F.S., Cruz, A.C.R., de Almeida Medeiros, D.B., da Silva, S.P., Nunes, M.R.T., Martins, L.C., Chiang, J.O., da Silva, L.P., Cunha, G.M., de Araujo, R.F., de Oliveira Monteiro, H.A. & Nunes Neto, J.P. (2020) Mitochondrial genome sequencing and phylogeny of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis (Diptera: Culicidae). Scientific Reports, 10, 16948. https://doi.org/10.1038/s41598-020-73790-x
- Deng, J.L., Yu, Z.Q., Ning, J., Wang, H., Lin, X.D. & Liu, X.L. (2020) Complete mitochondrial genome and phylogenetic analysis of a Chorthippus fallax (Zuboxsky) isolated in rangeland of northwest region of China. Mitochondrial DNA Part B, 5, 61–62. https://doi.org/10.1080/23802359.2019.1695549
- Ding, X.H., Fu, Y., Zhou, X., Yang, S.B., Cao, Y.M., Hou, F.X., Liu, X.L. & Sun, T. (2022) Complete mitogenome of Calliptamus barbarus Costa (Orthoptera: Acrididae) and its phylogeny in Acridoidea. Zootaxa, 5213 (4), 427–440. https://doi.org/10.11646/zootaxa.5213.4.6
- Du, Z., Hasegawa, H., Cooley, J.R., Simon, C., Yoshimura, J., Cai, W., Sota, T. & Li, H. (2019) Mitochondrial Genomics Reveals Shared Phylogeographic Patterns and Demographic History among Three Periodical Cicada Species Groups. Molecular Biology and Evolution, 36, 1187–1200. https://doi.org/10.1093/molbev/msz051
- Du, Z., Wu, Y., Chen, Z., Cao, L., Ishikawa, T., Kamitani, S., Sota, T., Song, F., Tian, L., Cai, W. & Li, H. (2021) Global Phylogeography and Invasion History of the Spotted Lanternfly Revealed by Mitochondrial Phylogenomics. Evolutionary Applications, 14, 915–930. https://doi.org/10.1111/eva.13170
- Eberhard, J.R. & Wright, T.F. (2016) Rearrangement and evolution of mitochondrial genomes in parrots. Molecular Phylogenetics and Evolution, 94, 34–36. https://doi.org/10.1016/j.ympev.2015.08.011
- Fang, X., Wang, X., Mao, B., Xiao, Y., Shen, M. & Fu, Y. (2023) Comparative mitogenome analyses of twelve non-biting flies and provide insights into the phylogeny of Chironomidae (Diptera: Culicomorpha). Scientific Reports, 13, 9200. https://doi.org/10.1038/s41598-023-36227-9
- Gissi, C., Iannelli, F. & Pesole, G. (2008) Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity, 101, 301–320. https://doi.org/10.1038/hdy.2008.62
- Hurst, L.D. (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends in Genetics, 18, 486–487. https://doi.org/10.1016/S0168-9525(02)02722-1
- Jia, W.Z., Yan, H.B., Guo, A.J., Zhu, X.Q., Wang, Y.C., Shi, W.G., Chen, H.T., Zhan, F. & Zhang, S.H. (2010) Complete mitochondrial genomes of Taeniamulticeps, T. hydatigena and T. pisiformis: Additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics, 11, 447. https://doi.org/10.1186/1471-2164-11-447
- Julio, R., Albert, F.M., Sánchez-DelBarrio Juan Carlos, Sara, G.R., Pablo, L., Ramos-Onsins, S.E. & Alejandro, S.G. (2017) Dnasp 6: dna sequence polymorphism analysis of large data sets. Molecular Biology & Evolution, 12, 12.
- Junqueira, A.C.M., Azeredo-Espin, A.M.L., Paulo, D.F., Marinho, M.A.T., Tomsho, L.P., Drautz-Moses, D.I., Purbojati, R.W., Ratan, A. & Schuster, S.C. (2016) Large-Scale Mitogenomics Enables Insights into Schizophora (Diptera) Radiation and Population Diversity. Scientific Reports, 6, 21762. https://doi.org/10.1038/srep21762
- Katoh, K. & Standley, D.M. (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution, 41, 353–358. https://doi.org/10.1007/BF01215182
- Kerpedjiev, P., Hammer, S. & Hofacker, I.L. (2015) Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics, 31, 3377–3379. https://doi.org/10.1093/bioinformatics/btv372
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35 (6), 1547–1549. https://doi.org/10.1093/molbev/msy096
- Li, H., Leavengood, J.M., Chapman, E.G., Burkhardt, D., Song, F., Jiang, P., Liu, J., Zhou, X. & Cai, W. (2017) Mitochondrial Phylogenomics of Hemiptera Reveals Adaptive Innovations Driving the Diversification of True Bugs. Proceedings of the Royal Society B-Biological Sciences, 284, 20171223. https://doi.org/10.1098/rspb.2017.1223
- Li, R., Wang, Y., Shu, X., Meng, L. & Li, B. (2020) Complete mitochondrial genomes of three Oxya grasshoppers (Orthoptera) and their implications for phylogenetic reconstruction. Genomics, 112 (1), 289–296. https://doi.org/10.1016/j.ygeno.2019.02.008
- Liu, Z.Q., Wang, Y.Q., Zhao, J., Shi, Y. & Zhu, X.P. (2005) The mitochondrial genome organization of the rice frog, Fejervarya limnocharis, (Amphibia: Anura): a new gene order in the vertebrate mtDNA. Gene, 346, 145–151. https://doi.org/10.1016/j.gene.2004.10.013
- Lowe, T.M. & Chan, P.P. (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research, 44 (W1), W54–W57. https://doi.org/10.1093/nar/gkw413
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
- Qian, Y.H., Wu, H.Y., Ji, X.Y., Yu, W.W. & Du, Y.Z. (2014) Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of plecoptera based on mitogenomes. PLoS ONE, 9, e86328. https://doi.org/10.1371/journal.pone.0086328
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
- Salvato, P., Simonato, M., Battisti, A. & Negrisolo, E. (2008) The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics, 9, 331. https://doi.org/10.1186/1471-2164-9-331
- Simon, C. (1991) Molecular systematics at the species boundary: Exploiting conserved and variable regions of the mitochondrial genome of animals via direct sequencing from amplified DNA. In: Hewitt, G.M., Johnston, A.W.B. & Young, J.P.W. (Eds.), Molecular techniques in taxonomy. Springer-Verlag, Berlin, pp. 33–71. https://doi.org/10.1007/978-3-642-83962-7_4
- Song, H., Mariño-Pérez, R., Woller, D. & Cigliano, M.M. (2018) Evolution, Diversification, and Biogeography of Grasshoppers (Orthoptera: Acrididae). Insect Systematics and Diversity, 2, 1–25. https://doi.org/10.1093/isd/ixy008
- Sun, S., Li, Q., Kong, L. & Yu, H. (2018) Multiple reversals of strand asymmetry in molluscs mitochondrial genomes, and consequences for phylogenetic inferences. Molecular Phylogenetics and Evolution, 118, 223–231. https://doi.org/10.1016/j.ympev.2017.10.009
- Villanueva, R.A.M. & Chen, Z.J. (2016) Ggplot2: elegant graphics for data analysis. 2nd Edition. Springer, New York, New York, 260 pp.
- Wang, L., Chen, J., Xue, X., Qin, G., Gao, Y., Li, K., Zhang, Y. & Li, X. (2023) Comparative analysis of mitogenomes among three species of grasshoppers (Orthoptera: Acridoidea: Gomphocerinae) and their phylogenetic implications. Peer Journal, 11, e16550. https://doi.org/10.7717/peerj.16550
- Woller, D.A., Fontana, P., Mariño-Pérez, R. & Song, H. (2014) Studies in Mexican grasshoppers: Liladownsia fraile, a new genus and species of Dactylotini (Acrididae: Melanoplinae) and an updated molecular phylogeny of Melanoplinae. Zootaxa, 3793 (4), 475–495. https://doi.org/10.11646/zootaxa.3793.4.6
- Wu, X., Li, Y., Zhang, H., Yan, L. & Wu, X.B. (2016) The complete mitochondrial genome of Microhyla pulchra (Amphibia: Anura, Microhylidae). Mitochondrial DNA Part A, 27, 40–41. https://doi.org/10.3109/19401736.2013.869685
- Xiao, B., Chen, W., Hu, C.C. & Jiang, G.F. (2012) Complete mitochondrial genome of the groundhopper Alulatettix yunnanensis (Insecta: Orthoptera: Tetrigoidea). Mitochondrial DNA, 23, 286–287. https://doi.org/10.3109/19401736.2012.674122
- Xie, Y., Zhang, Z., Niu, L., Wang, Q., Wang, C., Lan, J., Deng, J., Fu, Y., Nie, H. & Yan, N. (2011) The mitochondrial genome of Baylisascaris procyonis. PLoS ONE, 6, e27066. https://doi.org/10.1371/journal.pone.0027066
- Xu, D.L., Yu, T.H. & Zhang, Y.L. (2020) Characterization of the Complete Mitochondrial Genome of Drabescus ineffectus and Roxasellana stellata (Hemiptera: Cicadellidae: Deltocephalinae: Drabescini) and Their Phylogenetic Implications. Insects, 11, 534. https://doi.org/10.3390/insects11080534
- Yan, L., Pape, T., Elgar, M.A., Gao, Y. & Zhang, D. (2019) Evolutionary History of Stomach Bot Flies in the Light of Mitogenomics. Systematic Entomology, 44, 797–809. https://doi.org/10.1111/syen.12356
- Ye, F., Easy, R.H., King, S.D., Cone, D.K. & You, P. (2017) Comparative analyses within Gyrodactylus (Platyhelminthes: Monogenea) mitochondrial genomes and conserved polymerase chain reaction primers for gyrodactylid mitochondrial DNA. Journal of Fish Disease, 40, 541–555. https://doi.org/10.1111/jfd.12539
- Yuan, L., Liu, H., Ge, X., Yang, G., Xie, G. & Yang, Y. (2022) A Mitochondrial Genome Phylogeny of Cleridae (Coleoptera, Cleroidea). Insects, 13, 118. https://doi.org/10.3390/insects13020118
- Zhang, D., Gao, F., Jakovlic, I., Zou, H, Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348–355. https://doi.org/10.1111/1755-0998.13096
- Zhang, D., Yan, L., Zhang, M., Chu, H., Cao, J., Li, K., Hu, D. & Pape, T. (2016) Phylogenetic Inference of Calyptrates, with the First Mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). International Journal of Biological Sciences, 12, 489. https://doi.org/10.7150/ijbs.12148
- Zheng, Z.M. (1990) Three new locust species from Hengduanshan Range of China (Orthoptera: Acridoidea). Acta Zootaxonomica Sinica, 15 (2), 196–200.
- Zhi, Y.C., Liu, B., Han, G.F., Yin, H. & Zhang, D.C. (2016) The complete mitochondrial genome of Kingdonella bicollina (Orthoptera: Acridoidea: Catantopidae). Mitochondrial DNA Part A, 27 (1), 391–392. https://doi.org/10.3109/19401736.2014.896000
- Zhou, J. & Yang, D. (2022) Mitochondrial Genomes Provide New Phylogenetic and Evolutionary Insights into Psilidae (Diptera: Brachycera). Insects, 13, 518. https://doi.org/10.3390/insects13060518