Abstract
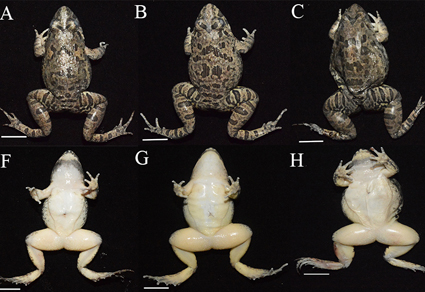
The Fejervarya limnocharis is a dicroglossid species complex distributed across east and southeast Asia, comprising four species. We previously found populations from Zhoushan Archipelago should be a distinct species with independent evolutionary history, through systematic classification based on mitochondrial DNA (mtDNA) and eight nuclear DNA loci, belonging to the fifth species in the complex. In this study, we described this new species as Fejervarya zhoushanensis sp. nov. by further comparing its genetic and morphological characteristics with its sister lineage F. kawamurai. The new species exhibited distinct genetic differences, as evidenced by genomic single nucleotide polymorphism (SNP) data. Morphological distinctions include larger snout-to-vent length (SVL), longer head length (HL), larger eye diameter (ED), greater distance between aeddistance anterior eye corners (AED), longer forearm length (FLL) and foot length (FL). Additionally, males exhibited larger tympanum diameter (TD), snout length (SL) and inner metatarsal tubercle length (IMTL); while females showed longer hindlimb length (HLL). These differences enabled clear taxonomic separation between F. zhoushanensis sp. nov. and F. kawamurai.
References
- Alexander, D.H., Novembre, J. & Lange, K. (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655–1664. https://doi.org/10.1101/gr.094052.109
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed 20 February 2025)
- Bolkay, S.J. (1915) Beiträge zur Osteologie einiger exotischer Raniden. Anatomischer Anzeiger, 48, 172–183. https://doi.org/10.5962/bhl.part.17101
- Bouckaert, R., Vaughan, T.G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., Heled, J., Jones, G., Kühnert, D., De Maio, N., Matschiner, M., Mendes, F.K., Müller, N.F., Ogilvie, H.A., du Plessis, L., Popinga, A., Rambaut, A., Rasmussen, D., Siveroni, I., Suchard, M.A., Wu, C.H., Xie, D., Zhang, C., Stadler, T. & Drummond, A.J. (2019) BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 15, e1006650. https://doi.org/10.1371/journal.pcbi.1006650
- Bouckaert, R.R. (2010) DensiTree: making sense of sets of phylogenetic trees. Bioinformatics, 26, 1372–1373. https://doi.org/10.1093/bioinformatics/btq110
- Brown, R.P., Jin, Y.T., Thomas, J. & Meloro, C. (2023) Life on a beach leads to phenotypic divergence despite gene flow for an island lizard. Communications Biology, 6, 141. https://doi.org/10.1038/s42003-023-04494-x
- Chan, K.O. & Grismer, L.L. (2021) A standardized and statistically defensible framework for quantitative morphological analyses in taxonomic studies. Zootaxa, 5023 (2), 293–300. https://doi.org/10.11646/zootaxa.5023.2.9
- Conix, S. (2018) Integrative taxonomy and the operationalization of evolutionary independence. European Journal for Philosophy of Science, 8, 587–603. https://doi.org/10.1007/s13194-018-0202-z
- Danecek, P., Auton, A., Abecasis, G., Albers, C.A., Banks, E., DePristo, M.A., Handsaker, R.E., Lunter, G., Marth, G.T., Sherry, S.T., McVean, G., Durbin, R. & Genomes Project Anal, G. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. https://doi.org/10.1093/bioinformatics/btr330
- Darriba, D., Posada, D., Kozlov, A.M., Stamatakis, A., Morel, B. & Flouri, T. (2020) ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution, 37, 291–294. https://doi.org/10.1093/molbev/msz189
- de Queiroz, K. (2007) Species concepts and species delimitation. Systematic Biology, 56, 879–886. https://doi.org/10.1080/10635150701701083
- Djong, H.T., Islam, M.M., Nishioka, M., Matsui, M., Ota, H., Kuramoto, M., Alam, M., Anslem, D. & Khonsue, W. (2007a) Genetic relationships and reproductive isolation mechanisms among the Fejervarya limnocharis complex from Indonesia (Java) and other Asian countries. Zoological Science, 24, 360–375. https://doi.org/10.2108/zsj.24.360
- Djong, H.T., Matsui, M., Kuramoto, M., Daicus, M.B., Yong, H.-S., Nishioka, M. & Sumida, M. (2007b) Morphological divergence, reproductive isolating mechanism and molecular phylogenetic relationship among Indonesia, Malaysia and Japan populations of the Fejervarya limnocharis complex (Anura, Ranidae). Zoological Science, 24, 1197–1212. https://doi.org/10.2108/zsj.24.1197
- Djong, H.T., Matsui, M., Kuramoto, M., Nishioka, M. & Sumida, M. (2011) A new species of the Fejervarya limnocharis complex from Japan (Anura, Dicroglossidae). Zoological Science, 28, 922–929. https://doi.org/10.2108/zsj.28.922
- Douglas, J. & Bouckaert, R. (2022) Quantitatively defining species boundaries with more efficiency and more biological realism. Communications Biology, 5, 755. https://doi.org/10.1038/s42003-022-03723-z
- Dubois, A. & Ohler, A. (2000) Systematics of Fejervarya limnocharis (Gravenhorst, 1829) (Amphibian, Anura, Ranidae) and related species. 1. Nomenclatural status and type‑specimens of the nominal species Rana limnocharis Gravenhorst, 1829. Alytes, 18, 15–50.
- Fei, L., Ye, C.Y., Jian, J.P. & Xie, F. (2002) On taxonomic status of Rana limnocharis group with revision of nomenclature of the rice frog from China. Herpetological Sinica, 9, 88–96.
- Frost, D.R. (2025) Amphibian species of the world. Version 6.2. An online reference. American Museum of Natural History, New York, New York. Available from: https://amphibiansoftheworld.amnh.org/ (accessed 20 February 2025)
- Futuyma, D.J. & Kirkpatrick, M. (2017) Evolution. Sinauer Associates, Inc., Sunderland.
- Hazkani-Covo, E., Zeller, R.M. & Martin, W. (2010) Molecular poltergeists: mitochondrial DNA copies (NUMTs) in Sequenced Nuclear genomes. PLoS Genetic, 6, e1000834. https://doi.org/10.1371/journal.pgen.1000834
- Jin, Y.T. & Brown, R.P. (2019) Morphological species and discordant mtDNA: A genomic analysis of Phrynocephalus lizard lineages on the Qinghai‑Tibetan Plateau. Molecular Phylogenetics and Evolution, 139, 106523. https://doi.org/10.1016/j.ympev.2019.106523
- Kotaki, M., Kurabayashi, A., Matsui, M., Kuramoto, M., Djong, T.H. & Sumida, M. (2010) Molecular phylogeny of the diversified frogs of genus Fejervarya (Anura: Dicroglossidae). Zoological Science, 27, 386–395. https://doi.org/10.2108/zsj.27.386
- Kozlov, A.M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. (2019) RAxML‑NG: a fast, scalable and user‑friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, 35, 4453–4455. https://doi.org/10.1093/bioinformatics/btz305
- Leaché, A.D., Fujita, M.K., Minin, V.N. & Bouckaert, R.R. (2014) Species delimitation using genome‑wide SNP data. Systematic Biology, 63, 534–542. https://doi.org/10.1093/sysbio/syu018
- Leese, F., Kop, A., Wägele, J.W. & Held, C. (2008) Cryptic speciation in a benthic isopod from Patagonian and Falkland Island waters and the impact of glaciations on its population structure. Frontiers in Zoology, 5, 19. https://doi.org/10.1186/1742-9994-5-19
- Letunic, I. & Bork, P. (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Research, 47, W256–W259. https://doi.org/10.1093/nar/gkz239
- Li, J.B. (2008) Regional geology of the east China sea. Ocean Press, Beijing, 631 pp.[in Chinese]
- Lin, L.H., Zhao, Q. & Ji, X. (2008) Conservation genetics of the Chinese cobra (Naja atra) investigated with mitochondrial DNA sequences. Zoological Science, 25, 888–893. https://doi.org/10.2108/zsj.25.888
- Marinho, P., Santos, M.T.T., Faivovich, J., Lyra, M.L., Giaretta, A.A., Haddad, C.F.B. & Carvalho, T.R. (2024) A new species of the Aplastodiscus albosignatus group (Hylinae: Cophomantini) from the northern Mantiqueira mountain range. Herpetologica, 80, 51–66. https://doi.org/10.1655/Herpetologica-D-23-00008
- Matsui, M., Toda, M. & Ota, H. (2007) A new species of frog allied to Fejervarya limnocharis from the Southern Ryukyus, Japan (Amphibia: Ranidae). Current Herpetology, 26, 65–79. https://doi.org/10.3105/1881-1019(2007)26[65:ANSOFA]2.0.CO;2
- Melo, A.T.O., Bartaula, R. & Hale, I. (2016) GBS‑SNP‑CROP: a reference‑optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired‑end genotyping‑by‑sequencing data. BMC Bioinformatics, 17, 29. https://doi.org/10.1186/s12859-016-0879-y
- Purcell, S., Neale, B., Todd‑Brown, K., Thomas, L., Ferreira, M.A.R., Bender, D., Maller, J., Sklar, P., de Bakker, P.I.W., Daly, M.J. & Sham, P.C. (2007) PLINK: A tool set for whole‑genome association and population‑based linkage analyses. The American Journal of Human Genetics, 81, 559–575. https://doi.org/10.1086/519795
- Quesada‑Ruiz, L.C. & Peña‑Alonso, C. (2023) Studies of environmental coastal impacts in small islands: a review. International Journal of Environment and Pollution, 72, 2–4. https://doi.org/10.1504/IJEP.2023.137972
- Rambaut, A. (2018) FigTree. Version 1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 20 February 2025)
- Schubert, M., Lindgreen, S. & Orlando, L. (2016) AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Research Notes, 9, 88. https://doi.org/10.1186/s13104-016-1900-2
- Silva, L.A., Marques, R., Folly, H. & Santana, D.J. (2022) A new Amazonian species of Allobates Zimmermann & Zimmermann, 1988 (Aromobatidae) with a trilled advertisement call. PeerJ, 10, e13026. https://doi.org/10.7717/peerj.13026
- Stoltz, M., Baeumer, B., Bouckaert, R., Fox, C., Hiscott, G. & Bryant, D. (2021) Bayesian inference of species trees using diffusion models. Systematic Biology, 70, 145–161. https://doi.org/10.1093/sysbio/syaa051
- Suárez, N.M., Pestano, J. & Brown, R.P. (2014) Ecological divergence combined with ancient allopatry in lizard populations from a small volcanic island. Molecular Ecology, 23, 4799–4812. https://doi.org/10.1111/mec.12897
- Tamar, K., Mitsi, P., Simó‑Riudalbas, M., Tejero‑Cicuéndez, H., Al‑Sariri, T. & Carranza, S. (2019) Systematics, biogeography, and evolution of Pristurus minimus (Squamata, Sphaerodactylidae) with the discovery of the smallest Arabian vertebrate. Systematics and Biodiversity, 17, 349–366. https://doi.org/10.1080/14772000.2019.1614694
- Toda, M., Nishida, M., Matsui, M., Wu, G.F. & Ota, H. (1997) Allozyme variation among east Asian populations of the Indian rice frog, Rana limnocharis (Amphibia: Anura). Biochemical Systematics and Ecology, 25, 143–159. https://doi.org/10.1016/S0305-1978(96)00098-1
- Toda, M., Matsui, M., Nishida, M. & Ota, H. (1998a) Genetic divergence among Southeast and East Asian populations of Rana limnocharis (Amphibia: Anura), with special reference to sympatric cryptic species in Java. Zoological Science, 15, 607–613. https://doi.org/10.2108/0289-0003(1998)15[607:GDASAE]2.0.CO;2
- Toda, M., Nishida, M., Matsui, M., Lue, K.Y. & Ota, H. (1998b) Genetic variation in the Indian rice frog, Rana limnocharis (Amphibia: Anura), in Taiwan, as revealed by allozyme data. Herpetologica, 54, 73–82.
- Tong, H.J., Wo, Y.B., Liao, P.H. & Jin, Y.T. (2017) Phylogenetic, demographic and dating analyses of Bufo gargarizans populations from the Zhoushan archipelago and mainland China. Asian Herpetological Research, 8, 165–173.
- Veith, M. (2001) Systematics of Fejervarya limnocharis (Gravenhorst, 1829) (Amphibia, Anura, Ranidae) and related species. 2. morphological and molecular variation in frogs from the Greater Sunda islands (Sumatra, Java, Borneo) with the definition of two species. Alytes, 25, 811–820.
- Wang, J. & Wang, P. (1982) Relationship between sea‑level changes and climatic fluctuation in East China since late Pleistocene. The Quaternary Research, 21, 101–114. https://doi.org/10.4116/jaqua.21.101
- Wei, L., Flanders, J.R., Rossiter, S.J., Miller‑Butterworth, C.M., Zhang, L.B. & Zhang, S.Y.Y. (2010) Phylogeography of the Japanese pipistrelle bat, Pipistrellus abramus, in China: the impact of ancient and recent events on population genetic structure. Biological Journal of the Linnean Society, 99, 582–594. https://doi.org/10.1111/j.1095-8312.2009.01387.x
- Xu, Q.Q. & Lin, H.M. (1993) An astroclimatological explanation of six marine transgressions in eastern China since middle Pleistocene. Marine Geology and Quaternary Geology, 13, 11–20. [in Chinese with English abstract]
- Yang, K., Wo, Y.B., Shao, G., Liao, P.H., Tong, H.J., Brown, R.P. & Jin, Y.T. (2022) Phylogenetic relationships among Chinese rice frogs within the Fejervarya limnocharis species complex (Amphibia: Dicroglossidae). Asian Herpetological Research, 13, 232–241.
- Zachos, F.E. (2018) Species concepts, species delimitation and the inherent limitations of taxonomy. Journal of Genetics, 97, 811–815. https://doi.org/10.1007/s12041-018-0965-1
- Zhong, J., Liu, Z.Q. & Wang, Y.Q. (2008) Phylogeography of the rice frog, Fejervarya multistriata (Anura: Ranidae), from China based on mtDNA D‑loop sequences. Zoological Science, 25, 811–820. https://doi.org/10.2108/zsj.25.811