Abstract
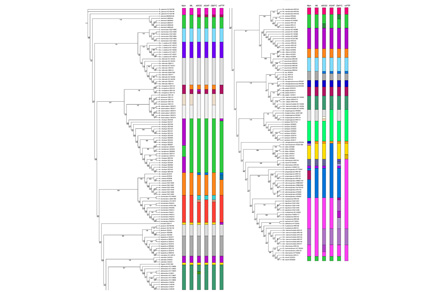
Grasshoppers are significant pests in agriculture, animal husbandry, and forestry. They are widely distributed and numerous species, with some taxa exhibiting considerable morphological variation, which poses substantial challenges for traditional taxonomic classification. DNA barcoding has become an essential tool for species identification, serving as an effective complement to morphological taxonomy. This study focuses on grasshoppers from the Bashang Grassland in Hebei Province, China. Mitochondrial cytochrome c oxidase subunit I (COI) gene sequences were used as DNA barcode sequences, which were obtained and analyzed using molecular biology techniques. In addition, homologous sequences of related species were retrieved from GenBank, resulting in a barcode dataset comprising 237 sequences from 42 species, 26 genera. Phylogenetic analysis barcoding gap assessment were performed, and multiple species delimitation methods—including ABGD, ASAP, GMYC, and mPTP—were applied for comprehensive evaluation. The results demonstrated that DNA barcoding can effectively delineate species-level taxonomic units for many grasshoppers, providing a powerful supplement to morphological taxonomy. However, the grasshopper taxa include both groups with high cryptic genetic diversity and closely related species that are difficult to distinguish using barcoding alone. Therefore, accurate identification of grasshopper species still requires integration with morphological approaches.
References
- Ahmad, J.N., Ahmad, S.J.N., Malik, M.A., Ali, A., Ali, M., Ahmad, E., Tahir, M. & Ashraf, M. (2020) Molecular Evidence for the Association of Swarm Forming Desert Locust, Schistocerca g. gregaria (Forskal) in Pakistan with Highly Prevalent Subspecies in Sahara Desert of Africa. Paskistan Journal Of Zoology, 52 (6), 2233–2242. https://doi.org/10.17582/journal.pjz/20200501020521
- Alvarado-Robledo, E.J., Hernández-Válazquez, I.M., Guillén-Navarro, K., Diego-García, E., Zarza, E. & Zamora Briseño, J.A. (2024) Metabarcoding: opportunities for accelerating monitoring and understanding insect tropical biodiversity. Journal Of Insect Conservation, 28 (4), 589–604. https://doi.org/10.1007/s10841-024-00584-1
- Chen, L.P., Chang, Y.G., Li, J., Wang, J.M., Liu, J.L., Zhi, Y.C. & Li, X.J. (2018) Application of DNA barcoding in the classification of grasshoppers (Orthoptera: Acridoidea) a case study of grasshoppers from Hebei Province, China. Zootaxa, 4497 (1), 99–110. https://doi.org/10.11646/zootaxa.4497.1.6
- Chen, S.L., Pang, X.H., Luo, K., Yao, H., Han, J.P. & Song, J.Y. (2013) DNA barcoding of biological resources. Chinese Bulletin of Life Sciences, 25 (5), 458–466.
- Decaëns, T., Porco, D., Rougerie, R., Brown, G.G. & James, S.W. (2013) Potential of DNA barcoding for earthworm research in taxonomy and ecology. Applied Soil Ecology, 65, 35–42. https://doi.org/10.1016/j.apsoil.2013.01.001
- Gernhard, T. (2008) The conditioned reconstructed process. Journal of Theoretical Biology, 253 (4), 769–778. https://doi.org/10.1016/j.jtbi.2008.04.005
- Guerrero, M.S. & Cayabyab, B.F. (2024) DNA barcoding of Locusta migratoria manilensis (Orthoptera: Acrididae) reveals insights into the species and subspecies differentiation. Journal Of Entomological Science, 59 (2), 125–132. https://doi.org/10.18474/JES23-36
- Hafayed, R., Moussi, A., Chang, H.H. & Huang, Y. (2023) Species delimitation and molecular phylogeny of the grasshopper subfamily Gomphocerinae (Orthoptera: Acrididae) from Algeria based on mitochondrial and nuclear DNA markers. African Zoology, 2023, 58 (3–4), 80–96. https://doi.org/10.1080/15627020.2023.2263498
- Hao, J.F., Zhang, X.H., Wang, Y.S., Liu, J.L., Zhi, Y.C. & Li, X.J. (2017) Diversity investigation and application of DNA barcoding of Acridoidea from Baiyangdian Wetland. Biodiversity Science, 25 (4), 409–417. https://doi.org/10.17520/biods.2016331
- Hao, J.Q. (2024) The occurrence of grassland pests in Hebei province in 2023 and the trend forecast in 2024. Journal of Hebei Forestry Science and Technology, (1), 65–66.
- Hawlitschek, O., Morinière, J., Lehmann, G.U.C., Lehmann, A.W., Kropf, M., Dunz, A., Glaw, F., Detcharoen, M., Schmidt, S., Hausmann, A., Szucsich, N.U., Caetano-Wyler, S.A. & Haszprunar, G. (2017) DNA barcoding of crickets, katydids and grasshoppers (Orthoptera) from Central Europe with focus on Austria, Germany and Switzerland. Molecular Ecology Resources, 17 (5), 1037–1053. https://doi.org/10.1111/1755-0998.12638
- Hebert, P.D.N., Cywinska, A., Ball, S.L. & DeWaard, J.R. (2003) Biological identifications through DNA barcodes. The Royal Society Proceedings B, 270, 313–321. https://doi.org/10.1098/rspb.2002.2218
- Hebert, P.D.N. & Gregory, T.R. (2005) The promise of DNA barcoding for taxonomy. Systematic Biology, 54, 852–859. https://doi.org/10.1080/10635150500354886
- Huang, J.H., Zhang, A.B., Mao, S.L. & Huang, Y. (2013) DNA barcoding and species boundary delimitation of selected species of Chinese Acridoidea (Orthoptera: Caelifera). PLoS One, 8 (12), e82400. https://doi.org/10.1371/journal.pone.0082400
- Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A. & Flouri, T. (2017) Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics, 33 (11), 1630–1638. https://doi.org/10.1093/bioinformatics/btx025
- Mo, W.H., Chen, H.Y., Pang, H. & Liu, J.X. (2021) DNA barcoding for molecular identification of the genus Oxyscelio (Hymenoptera: Scelionidae) from southern China, with descriptions of five new species. Journal of Hymenoptera Research, 87, 613–633. https://doi.org/10.3897/jhr.87.71912
- Nguyen, L.T., Schmidt, H.A., von Haesele, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32 (1), 268–274. https://doi.org/10.1093/molbev/msu300
- Pan, C.Y., Hu, J., Zhang, X. & Huang, Y. (2006) The DNA Barcoding Application of mtDNA COⅠ Genes in Seven Species of Catantopidae (Orthoptera). Entomotaxonomia, 28 (2), 103–110.
- Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. (2012) ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology, 21 (8), 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
- Puillandre, N., Brouillet, S. & Achaz, G. (2021) ASAP: assemble species by automatic partitioning. Molecular ecology resources, 21 (2), 609–620. https://doi.org/10.1111/1755-0998.13281
- Ronquist, F., Teslenko, M., van der Mark, P., Daniel, L., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61 (3), 539–542. https://doi.org/10.1093/sysbio/sys029
- Rozewicki, J., Li, S.L., Karlou Mar Amada, D.M. Standley & Katoh, K. (2019) MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Research, 47 (W1), 5–10. https://doi.org/10.1093/nar/gkz342
- Suchard, M.A., Lemey, P., Baele, G., Ayres, D.L., Drummond, A.J. & Rambaut, A. (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4 (1), vey016. https://doi.org/10.1093/ve/vey016
- Talavera, G. & Castresana, J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56 (4), 564–577. https://doi.org/10.1080/10635150701472164
- Velzen, R.V., Weitschek, E., Felici, G. & Bakker, F.T. (2012) DNA barcoding of recently diverged species: relative performance of matching methods [J]. PLoS One, 7 (1), e30490. https://doi.org/10.1371/journal.pone.0030490
- Wang, L., Chen, J.Y., Xue, X.B., Qin, G.Q., Gao, Y.Y., Li, K., Zhang, Y.L. & Li, X.J. (2023) Comparative analysis of mitogenomes among three species of grasshoppers (Orthoptera: Acridoidea: Gomphocerinae) and their phylogenetic implications. Peer J, 11, e16550. https://doi.org/10.7717/peerj.16550
- Xu, M.Y., Li, Y.Q., Wang, K., Cao, Y.F., Yu, H.L., Li, X.F., Li, L.S., X., Jing, F.T. & Xie, F. (2009) Spatial distribution and dynamic characteristics of the grassland vegetation in Hebei. Acta Prataculturae Sinica, 18 (6), 1–11.
- Yang, Z.M., Li, J.Z., Zhang, H.Q., Wang, Y.T., Liu, J.C., Huang, J.S., Li, F., Dong, X.F., Wang, W.J., Zhang, J.P., Li, G.Y., Wang, W.T. & Tong, W.H. (2021) The Analysis of the species diversity and fauna distribution of grasshoppers in Bashang Area of Hebei Province [J]. Journal of Anhui Agricultural Sciences, 49 (22), 143–147.
- Zhang, D., Gao, F.L., Jakovlić, I., Zou, H., Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20 (1), 348–355. https://doi.org/10.1111/1755-0998.13096
- Zhang, P., Cai, Y., Ma, L., Chai, J. & Zhou, Z. (2024) DNA barcoding of the genus Gampsocleis (Orthoptera: Tettigoniidae) from China. Archives of Insect Biochemistry and Physiology, 115 (1), e22070. https://doi.org/10.1002/arch.22070
- Zhao, L., Zhao, L., Li, Q. & Ren, J.L. (2015) DNA barcoding of nymph grasshoppers from Xinjiang [J]. Xinjiang Agricultural Sciences, 52 (5), 785–794.